2RHZ
 
 | |
2Z34
 
 | |
2Z3F
 
 | |
5BO0
 
 | |
5DX0
 
 | |
5DWQ
 
 | |
7V1L
 
 | |
1NG0
 
 | |
6WK2
 
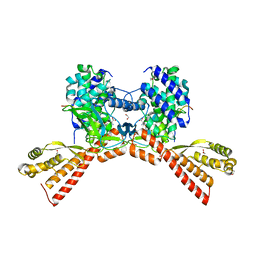 | | SETD3 mutant (N255V) in Complex with an Actin Peptide with His73 Replaced with Methionine | | Descriptor: | 1,2-ETHANEDIOL, Actin, cytoplasmic 2, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of SETD3 methyltransferase-mediated protein methionine methylation.
J.Biol.Chem., 295, 2020
|
|
6WK1
 
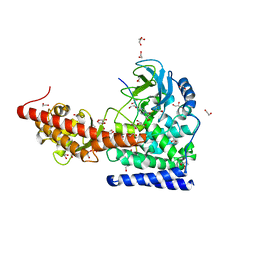 | | SETD3 in Complex with an Actin Peptide with His73 Replaced with Methionine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization of SETD3 methyltransferase-mediated protein methionine methylation.
J.Biol.Chem., 295, 2020
|
|
5V11
 
 | |
1OOJ
 
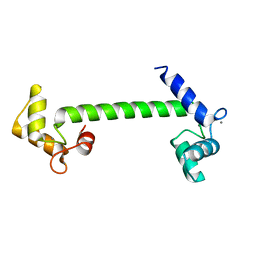 | | Structural genomics of Caenorhabditis elegans : Calmodulin | | Descriptor: | CALCIUM ION, Calmodulin CMD-1 | | Authors: | Symersky, J, Lin, G, Li, S, Qiu, S, Luan, C.-H, Luo, D, Tsao, J, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural genomics of caenorhabditis elegans: crystal structure of calmodulin.
Proteins, 53, 2003
|
|
5H7D
 
 | | Crystal structure of the YgjG-protein A-Zpa963-calmodulin complex | | Descriptor: | CALCIUM ION, Putrescine aminotransferase,Immunoglobulin G-binding protein A, Zpa963,Calmodulin | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
8SR6
 
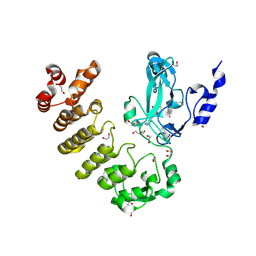 | | Crystal structure of legAS4 from Legionella pneumophila subsp. pneumophila with histone H3 (3-17)peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Eukaryotic huntingtin interacting protein B, ... | | Authors: | Xu, C, Chung, I.Y.W, Cygler, M. | | Deposit date: | 2023-05-05 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The SET and ankyrin domains of the secreted Legionella pneumophila histone methyltransferase work together to modify host chromatin.
Mbio, 14, 2023
|
|
2KNT
 
 | |
6G0L
 
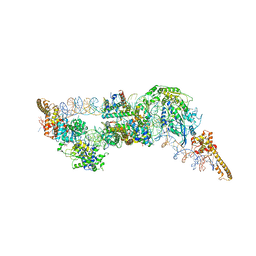 | | Structure of two molecules of the chromatin remodelling enzyme Chd1 bound to a nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
6ITU
 
 | | Crystal Structure of the GULP1 PTB domain-APP peptide complex | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 protein, PTB domain-containing engulfment adapter protein 1, ... | | Authors: | Yung, K.W.Y, Lau, K.F, Ngo, J.C.K. | | Deposit date: | 2018-11-26 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Attenuation of amyloid-beta generation by atypical protein kinase C-mediated phosphorylation of engulfment adaptor PTB domain containing 1 threonine 35.
Faseb J., 33, 2019
|
|
7DBH
 
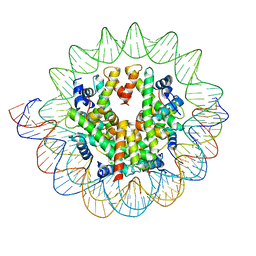 | | The mouse nucleosome structure containing H3mm18 | | Descriptor: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2020-10-20 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
7VBM
 
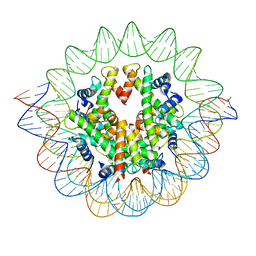 | | The mouse nucleosome structure containing H3mm18 aided by PL2-6 scFv | | Descriptor: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2021-08-31 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
