2ATL
 
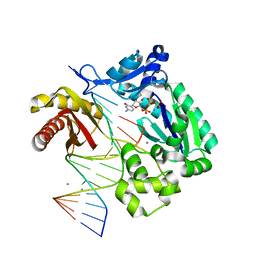 | | Unmodified Insertion Ternary Complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*T*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2005-08-25 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
4KQ0
 
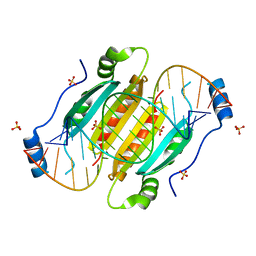 | | Crystal structure of double-helical CGG-repetitive RNA 19mer complexed with RSS p19 | | Descriptor: | 5'-R(P*GP*GP*CP*GP*GP*CP*GP*GP*CP*GP*GP*CP*GP*GP*CP*GP*GP*CP*C)-3', RNA silencing suppressor p19, SULFATE ION | | Authors: | Cabo, A, Katorcha, E, Tamjar, J, Popov, A.N, Malinina, L. | | Deposit date: | 2013-05-14 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into CNG-repetitive RNAs associated with human Trinucleotide Repeat Expansion Diseases (TREDs)
To be Published
|
|
2DRU
 
 | | Crystal structure and binding properties of the CD2 and CD244 (2B4) binding protein, CD48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, chimera of CD48 antigen and T-cell surface antigen CD2 | | Authors: | Evans, E.J, Ikemizu, S, Davis, S.J. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Binding Properties of the CD2 and CD244 (2B4)-binding Protein, CD48
J.Biol.Chem., 281, 2006
|
|
3GRS
 
 | |
2GHP
 
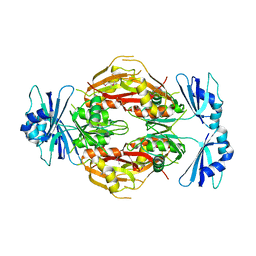 | | Crystal structure of the N-terminal 3 RNA binding domains of the yeast splicing factor Prp24 | | Descriptor: | U4/U6 snRNA-associated splicing factor PRP24 | | Authors: | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and interactions of the first three RNA recognition motifs of splicing factor prp24.
J.Mol.Biol., 367, 2007
|
|
2GPA
 
 | | ALLOSTERIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Tsitsanou, K.E, Zographos, S.E, Skamnaki, V.T. | | Deposit date: | 1999-02-18 | | Release date: | 1999-02-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric inhibition of glycogen phosphorylase a by the potential antidiabetic drug 3-isopropyl 4-(2-chlorophenyl)-1,4-dihydro-1-ethyl-2-methyl-pyridine-3,5,6-tricarbo xylate.
Protein Sci., 8, 1999
|
|
1WEJ
 
 | | IGG1 FAB FRAGMENT (OF E8 ANTIBODY) COMPLEXED WITH HORSE CYTOCHROME C AT 1.8 A RESOLUTION | | Descriptor: | CYTOCHROME C, E8 ANTIBODY, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mylvaganam, S.E, Paterson, Y, Getzoff, E.D. | | Deposit date: | 1998-03-26 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of an anti-cytochrome c antibody to its antigen: crystal structures of FabE8-cytochrome c complex to 1.8 A resolution and FabE8 to 2.26 A resolution.
J.Mol.Biol., 281, 1998
|
|
2K4F
 
 | | Mouse CD3epsilon Cytoplasmic Tail | | Descriptor: | T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Xu, C, Call, M.E, Schwieters, C.D, Schnell, J.R, Gagnon, E.E, Wucherpfennig, K.W, Chou, J.J. | | Deposit date: | 2008-06-07 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Regulation of T Cell Receptor Activation by Dynamic Membrane Binding of the CD3varepsilon Cytoplasmic Tyrosine-Based Motif.
Cell(Cambridge,Mass.), 135, 2008
|
|
2ATI
 
 | | Glycogen Phosphorylase Inhibitors | | Descriptor: | Glycogen phosphorylase, liver form, N-(2-CHLORO-4-FLUOROBENZOYL)-N'-(5-HYDROXY-2-METHOXYPHENYL)UREA, ... | | Authors: | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.J, Herling, A.W, Oikonomakos, N.G, Schmoll, D, Sarubbi, E, von Roedern, E, Schoenafinger, K, Defossa, E. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acyl ureas as human liver glycogen phosphorylase inhibitors for the treatment of type 2 diabetes.
J.Med.Chem., 48, 2005
|
|
2JWA
 
 | |
2ASD
 
 | | oxoG-modified Insertion Ternary Complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*T*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2005-08-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
1WW2
 
 | | Crystallographic studies on two bioisosteric analogues, N-acetyl-beta-D-glucopyranosylamine and N-trifluoroacetyl-beta-D-glucopyranosylamine, potent inhibitors of muscle glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, muscle form, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Anagnostou, E, Kosmopoulou, M.N, Chrysina, E.D, Leonidas, D.D, Hadjiloi, T, Tiraidis, C, Zographos, S.E, Gyorgydeak, Z, Somsak, L, Docsa, T, Gergely, P, Kolisis, F.N, Oikonomakos, N.G. | | Deposit date: | 2004-12-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies on two bioisosteric analogues, N-acetyl-beta-d-glucopyranosylamine and N-trifluoroacetyl-beta-d-glucopyranosylamine, potent inhibitors of muscle glycogen phosphorylase
Bioorg.Med.Chem., 14, 2006
|
|
1NIW
 
 | | Crystal structure of endothelial nitric oxide synthase peptide bound to calmodulin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nitric-oxide synthase, ... | | Authors: | Aoyagi, M, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-12-26 | | Release date: | 2003-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for endothelial nitric oxide synthase binding to calmodulin
Embo J., 22, 2003
|
|
1YYK
 
 | | Crystal structure of RNase III from Aquifex Aeolicus complexed with double-stranded RNA at 2.5-angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YYO
 
 | | Crystal structure of RNase III mutant E110K from Aquifex aeolicus complexed with double-stranded RNA at 2.9-Angstrom Resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1E2T
 
 | | Arylamine N-acetyltransferase (NAT) from Salmonella typhimurium | | Descriptor: | N-HYDROXYARYLAMINE O-ACETYLTRANSFERASE | | Authors: | Sinclair, J.C, Sandy, J, Delgoda, R, Sim, E, Noble, M.E.M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-07-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Arylamine N-Acetyltransferase Reveals a Catalytic Triad
Nat.Struct.Biol., 7, 2000
|
|
2H5U
 
 | | Crystal structure of laccase from Cerrena maxima at 1.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lyashenko, A.V, Gabdoulkhakov, A.G, Zaitsev, V.N, Lamzin, V.S, Lindley, P.F, Bento, I, Betzel, C, Zhukhlistova, N.E, Zhukova, Y.N, Mikhailov, A.M. | | Deposit date: | 2006-05-27 | | Release date: | 2007-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Purification, crystallization and preliminary X-ray study of the fungal laccase from Cerrena maxima
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2KPE
 
 | | Refined structure of Glycophorin A transmembrane segment dimer in DPC micelles | | Descriptor: | Glycophorin-A | | Authors: | Mineev, K.S, Bocharov, E.V, Goncharuk, M.V, Arseniev, A.S, Volynsky, P.E, Efremov, R.G. | | Deposit date: | 2009-10-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dimeric structure of the transmembrane domain of glycophorin a in lipidic and detergent environments.
Acta Naturae, 3, 2011
|
|
1EJC
 
 | | Crystal structure of unliganded mura (type2) | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
8PZQ
 
 | | Model for focused reconstruction of influenza A RNP-like particle | | Descriptor: | Nucleoprotein, RNA (5'P-(UC)6-FAM3') | | Authors: | Chenavier, F, Estrozi, L.F, Zarkadas, E, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation.
Sci Adv, 9, 2023
|
|
4X5C
 
 | | Anthranilate phosphoribosyltransferase variant R193L from Mycobacterium tuberculosis with pyrophosphate/PRPP and Mg2+ bound | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Cookson, T.V.M, Parker, E.J, Lott, J.S. | | Deposit date: | 2014-12-05 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structures of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Variants Reveal the Conformational Changes That Facilitate Delivery of the Substrate to the Active Site.
Biochemistry, 54, 2015
|
|
3HSZ
 
 | | Crystal structure of E. coli HPPK(F123A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pterin-binding site mutation Y53A, N55A or F123A and activity of E. coli HPPK
To be Published
|
|
6Y4J
 
 | | Engineered Fructosyl Peptide Oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase, GLYCEROL, ... | | Authors: | Donini, S, Rigoldi, F, Gautieri, A, Parisini, E. | | Deposit date: | 2020-02-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Rational backbone redesign of a fructosyl peptide oxidase to widen its active site access tunnel.
Biotechnol.Bioeng., 117, 2020
|
|
6Y8N
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with Redx03863 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1~{S},5~{R})-6-azanyl-3-azabicyclo[3.1.0]hexan-3-yl]-6-fluoranyl-~{N}-methyl-2-(2-methylpyrimidin-5-yl)oxy-9~{H}-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
2VIG
 
 | | Crystal structure of human short-chain acyl CoA dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A PERSULFIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pike, A.C.W, Pantic, N, Parizotto, E, Gileadi, O, Ugochukwu, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Short-Chain Acyl Coa Dehydrogenase
To be Published
|
|
