6JQN
 
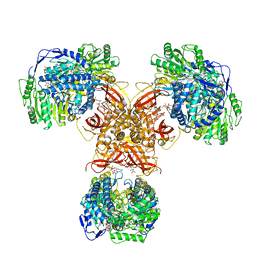 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and OCoA | | Descriptor: | Bifunctional protein PaaZ, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQO
 
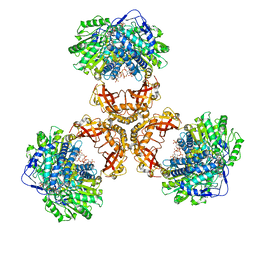 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and CCoA | | Descriptor: | Bifunctional protein PaaZ, CROTONYL COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQL
 
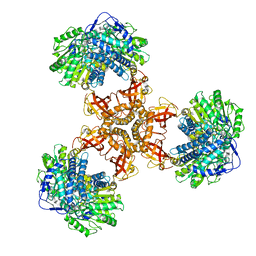 | | Structure of PaaZ, a bifunctional enzyme | | Descriptor: | Bifunctional protein PaaZ | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQM
 
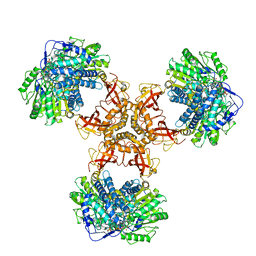 | | Structure of PaaZ with NADPH | | Descriptor: | Bifunctional protein PaaZ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
3P9T
 
 | | SmeT-Triclosan complex | | Descriptor: | Repressor, SULFATE ION, TRICLOSAN | | Authors: | Hernandez, A, Ruiz, F.M, Romero, A, Martinez, J.L. | | Deposit date: | 2010-10-18 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Binding of Triclosan to SmeT, the Repressor of the Multidrug Efflux Pump SmeDEF, Induces Antibiotic Resistance in Stenotrophomonas maltophilia.
Plos Pathog., 7, 2011
|
|
6UEH
 
 | | Crystal structure of a ruminal GH26 endo-beta-1,4-mannanase | | Descriptor: | ACETATE ION, CALCIUM ION, Cow rumen GH26 endo-mannanase | | Authors: | Mandelli, F, Morais, M.A.B, Lima, E.A, Persinoti, G.F, Murakami, M.T. | | Deposit date: | 2019-09-21 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Spatially remote motifs cooperatively affect substrate preference of a ruminal GH26-type endo-beta-1,4-mannanase.
J.Biol.Chem., 295, 2020
|
|
6T15
 
 | | The III2-IV(5B)1 respiratory supercomplex from S. cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CARDIOLIPIN, COPPER (II) ION, ... | | Authors: | Marechal, A, Pinotsis, N, Hartley, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Rcf2 revealed in cryo-EM structures of hypoxic isoforms of mature mitochondrial III-IV supercomplexes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8RY2
 
 | |
4G8N
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist G8M | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]cyclobutanecarboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Kastrup, J.S, Frydenvang, K, Gajhede, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors.
J.Struct.Biol., 180, 2012
|
|
7EY3
 
 | | Double cysteine mutations in T1 lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Hamdan, S.H, Leow, T.C, Yahaya, N.M, Ali, M.S.M, Jonet, M.A, Mohamad Aris, S.N.A, Maiangwa, J. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Knotting terminal ends of mutant T1 lipase with disulfide bond improved structure rigidity and stability.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
3SNO
 
 | |
2D5R
 
 | | Crystal Structure of a Tob-hCaf1 Complex | | Descriptor: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | Authors: | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | Deposit date: | 2005-11-04 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
5ZIN
 
 | | Crystal structure of bacteriorhodopsin at 1.27 A resolution | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Hasegawa, N, Jonotsuka, H, Miki, K, Takeda, K. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | X-ray structure analysis of bacteriorhodopsin at 1.3 angstrom resolution.
Sci Rep, 8, 2018
|
|
3GIP
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc, acetate and formate ions. | | Descriptor: | ACETIC ACID, FORMIC ACID, N-acyl-D-glutamate deacylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3GIQ
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc and phosphonate inhibitor, a mimic of the reaction tetrahedral intermediate. | | Descriptor: | N-[(R)-hydroxy(methyl)phosphoryl]-D-glutamic acid, N-acyl-D-glutamate deacylase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
5ZIL
 
 | | Crystal structure of bacteriorhodopsin at 1.29 A resolution | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Hasegawa, N, Jonotsuka, H, Miki, K, Takeda, K. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | X-ray structure analysis of bacteriorhodopsin at 1.3 angstrom resolution.
Sci Rep, 8, 2018
|
|
5ZIM
 
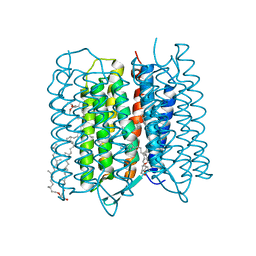 | | Crystal structure of bacteriorhodopsin at 1.25 A resolution | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Hasegawa, N, Jonotsuka, H, Miki, K, Takeda, K. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | X-ray structure analysis of bacteriorhodopsin at 1.3 angstrom resolution.
Sci Rep, 8, 2018
|
|
4INN
 
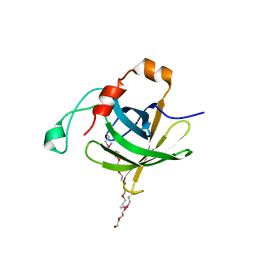 | | Protein HP1028 from the human pathogen Helicobacter pylori belongs to the lipocalin family | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, HP1028 | | Authors: | Barison, N, Cendron, L, Loconte, V, Zanotti, G. | | Deposit date: | 2013-01-05 | | Release date: | 2013-07-31 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein HP1028 from the human pathogen Helicobacter pylori belongs to the lipocalin family.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KZS
 
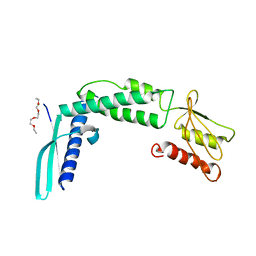 | | Crystal structure of the secreted protein HP1454 from the human pathogen Helicobacter pylori | | Descriptor: | LPP20 lipofamily protein, PENTAETHYLENE GLYCOL | | Authors: | Quarantini, S, Cendron, L, Fischer, W, Zanotti, G. | | Deposit date: | 2013-05-30 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the secreted protein HP1454 from the human pathogen Helicobacter pylori.
Proteins, 82, 2014
|
|
8VY4
 
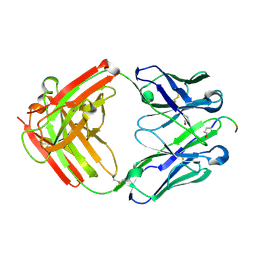 | | Engineering a Tumor-Selective Prodrug T Cell Engager Bispecific Antibody for Safer Immunotherapy | | Descriptor: | Fab Heavy Chain, Fab Light Chain, PCA-ASP-GLY-ASN-GLU-GLU-MET | | Authors: | Antonysamy, S, Demarest, S, Froning, K, Hickey, M.H, Kuhlman, B, McCue, A.C. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering a tumor-selective prodrug T-cell engager bispecific antibody for safer immunotherapy.
Mabs, 16, 2024
|
|
4OW6
 
 | | Crystal structure of Diphtheria Toxin at acidic pH | | Descriptor: | Diphtheria toxin | | Authors: | Leka, O, Vallese, F, Pirazzini, M, Berto, P, Montecucco, C, Zanotti, G. | | Deposit date: | 2014-01-31 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diphtheria toxin conformational switching at acidic pH.
Febs J., 281, 2014
|
|
1H5X
 
 | |
3OEZ
 
 | | crystal structure of the L317I mutant of the chicken c-Src tyrosine kinase domain complexed with imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, ACETATE ION, GLYCEROL, ... | | Authors: | Boubeva, R, Pernot, L, Perozzo, R, Scapozza, L. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | a single amino-acid dictates the dynamics of the switch between active and inactive C-Src conformation
To be Published
|
|
4F61
 
 | | Tubulin:Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gigant, B, Mignot, I, Knossow, M. | | Deposit date: | 2012-05-14 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Design and characterization of modular scaffolds for tubulin assembly.
J.Biol.Chem., 287, 2012
|
|
4F6R
 
 | | Tubulin:Stathmin-like domain complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Designed ankyrin repeat protein (DARPIN) D2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gigant, B, Mignot, I, Knossow, M. | | Deposit date: | 2012-05-15 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Design and characterization of modular scaffolds for tubulin assembly.
J.Biol.Chem., 287, 2012
|
|
