3VBB
 
 | |
4GJJ
 
 | |
3VSS
 
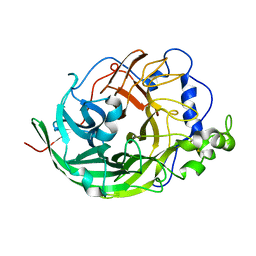 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain complexed with fructose | | Descriptor: | Beta-fructofuranosidase, beta-D-fructofuranose | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3VJB
 
 | |
6P8W
 
 | |
3VTA
 
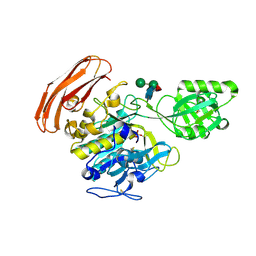 | | Crystal Structure of cucumisin, a subtilisin-like endoprotease from Cucumis melo L | | Descriptor: | Cucumisin, DIISOPROPYL PHOSPHONATE, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Murayama, K, Kato-Murayama, M, Hosaka, T, Sotokawauchi, A, Shirouzu, M, Arima, K, Yokoyama, S. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of cucumisin, a subtilisin-like endoprotease from Cucumis melo L
J.Mol.Biol., 423, 2012
|
|
4G01
 
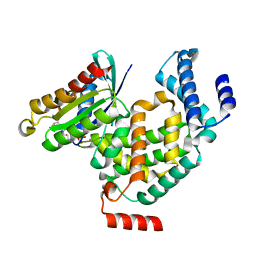 | | ARA7-GDP-Ca2+/VPS9a | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein RABF2b, ... | | Authors: | Ihara, K. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct metal recognition by guanine nucleotide-exchange factor in the initial step of the exchange reaction
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4UGP
 
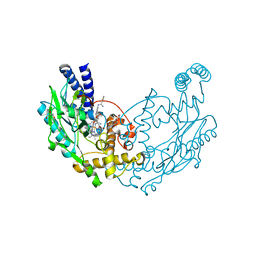 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with N',N'-(((2R)-3-aminopropane-1,2-diyl)bis(oxymethanediylbenzene-3,1- diyl))dithiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
6PDK
 
 | |
4UKD
 
 | | UMP/CMP KINASE FROM SLIME MOLD COMPLEXED WITH ADP, UDP, BERYLLIUM FLUORIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM DIFLUORIDE, MAGNESIUM ION, ... | | Authors: | Schlichting, I, Reinstein, J. | | Deposit date: | 1997-05-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of active conformations of UMP kinase from Dictyostelium discoideum suggest phosphoryl transfer is associative.
Biochemistry, 36, 1997
|
|
6P2N
 
 | | Crystal structure of Paenibacillus graminis GH74 (PgGH74) | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Stogios, P.J. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-31 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
4UIB
 
 | | Crystal structure of 3p in complex with tafCPB | | Descriptor: | (2S)-6-AMINO-2-[[(1R)-1-(CYCLOHEXYLMETHYL)-2-OXO-2-[[(2S)-1,7,7-TRIMETHYLNORBORNAN-2-YL]AMINO]ETHYL]ARBAMOYLAMINO]HEXANOIC ACID, ACETATE ION, CARBOXYPEPTIDASE B, ... | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
4UIA
 
 | | Crystal structure of 3a in complex with tafCPB | | Descriptor: | (2S)-6-azanyl-2-[[(2R)-3-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-propan-2-yl]carbamoylamino]hexanoic acid, CARBOXYPEPTIDASE B, ZINC ION | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
4UF6
 
 | | UCH-L5 in complex with ubiquitin-propargyl bound to an activating fragment of INO80G | | Descriptor: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, POLYUBIQUITIN-B, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UH0
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with N1-(6-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)pyridin-2- yl)-N1,N2-dimethylethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N'-[6-[2-(6-AZANYL-4-METHYL-PYRIDIN-2-YL)ETHYL]PYRIDIN-2-YL]-N,N'-DIMETHYL-ETHANE-1,2-DIAMINE, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-03-23 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | 2-Aminopyridines with a Truncated Side Chain to Improve Human Neuronal Nitric Oxide Synthase Inhibitory Potency and Selectivity.
J.Med.Chem., 58, 2015
|
|
4UDY
 
 | | NCO- bound to cluster C of Ni,Fe-CO dehydrogenase at true-atomic resolution | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Fesseler, J, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2014-12-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | How the [Nife4 S4 ] Cluster of Co Dehydrogenase Activates Co2 and Nco(.)
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4UHI
 
 | | HUMAN STEROL 14-ALPHA DEMETHYLASE (CYP51) IN COMPLEX WITH VFV IN C121 SPACE GROUP | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, I Lepesheva, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Human Sterol 14Alpha-Demethylase (Cyp51) as a Target for Anticancer Chemotherapy: Towards Structure-Aided Drug Design.
J.Lipid Res., 57, 2016
|
|
4UI6
 
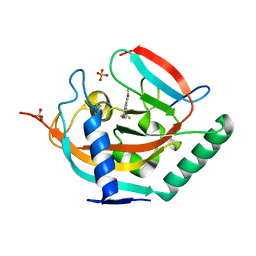 | | Crystal structure of human tankyrase 2 in complex with TA-47 | | Descriptor: | 8-METHOXY-2-[4-(TRIFLUOROMETHYL)PHENYL]-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4UM2
 
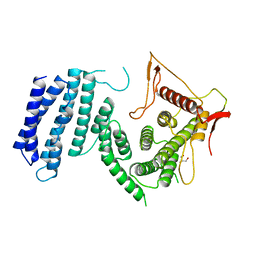 | | Crystal structure of the TPR domain of SMG6 | | Descriptor: | GLYCEROL, TELOMERASE-BINDING PROTEIN EST1A | | Authors: | Chakrabarti, S, Bonneau, F, Schuessler, S, Eppinger, E, Conti, E. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phospho-Dependent and Phospho-Independent Interactions of the Helicase Upf1 with the Nmd Factors Smg5-Smg7 and Smg6.
Nucleic Acids Res., 42, 2014
|
|
4V0C
 
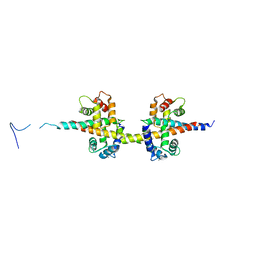 | |
6P48
 
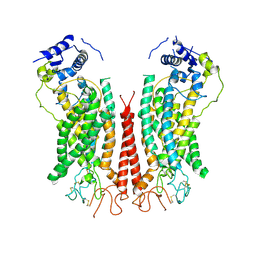 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl1 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
4UIW
 
 | | BROMODOMAIN OF HUMAN BRD9 WITH N-(1,1-dioxo-1-thian-4-yl)-5-ethyl-4- oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2- carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 9, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
4GC0
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | Descriptor: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4ULW
 
 | |
4UMT
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 1-(4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}-2-methoxybenzyl)piperazinediium, DIMETHYL SULFOXIDE, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
