4Q0U
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
6N4K
 
 | |
4P9E
 
 | |
4PN2
 
 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylotriose | | Descriptor: | CALCIUM ION, Xylanase, beta-D-xylopyranose | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4P9W
 
 | | Structure of ConA/Rh3Man | | Descriptor: | 2-[2-(2-{4-[(alpha-D-mannopyranosyloxy)methyl]-1H-1,2,3-triazol-1-yl}ethoxy)ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, CALCIUM ION, Concanavalin-A, ... | | Authors: | Sakai, F, Weiss, M.S, Jiang, M. | | Deposit date: | 2014-04-06 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of ConA/Rh3Man
To Be Published
|
|
4PAE
 
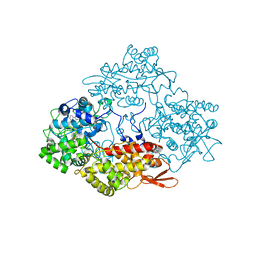 | | Crystal structure of catalase-peroxidase (KatG) W78F mutant from Synechococcus elongatus PCC7942 | | Descriptor: | Catalase-peroxidase, HEME B/C, SODIUM ION, ... | | Authors: | Kamachi, S, Wada, K, Tada, T. | | Deposit date: | 2014-04-08 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Crystal structure of the catalase-peroxidase KatG W78F mutant from Synechococcus elongatus PCC7942 in complex with the antitubercular pro-drug isoniazid.
Febs Lett., 589, 2015
|
|
4POO
 
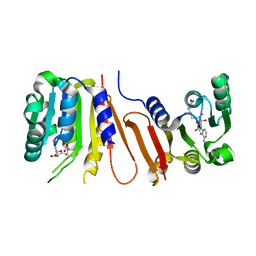 | | The crystal structure of Bacillus subtilis YtqB in complex with SAM | | Descriptor: | Putative RNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Park, S.C, Song, W.S, Yoon, S.I. | | Deposit date: | 2014-02-26 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of a putative SAM-dependent methyltransferase, YtqB, from Bacillus subtilis
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4PC0
 
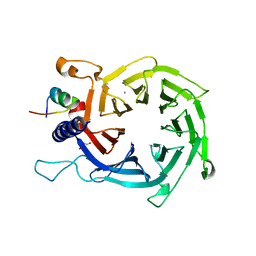 | | Structure of the human RbAp48-MTA1(670-711) complex | | Descriptor: | CALCIUM ION, GLYCEROL, Histone-binding protein RBBP4, ... | | Authors: | Alqarni, S.S.M, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
5U75
 
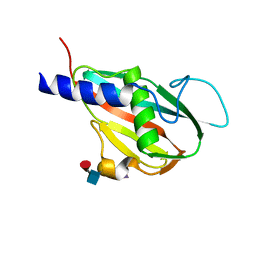 | | The structure of Staphylococcal Enterotoxin-like X (SElX), a Unique Superantigen | | Descriptor: | Enterotoxin-like toxin X, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Ting, Y.T, Young, P.G, Langley, R.J, Baker, H, Fraser, J.D. | | Deposit date: | 2016-12-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Staphylococcal enterotoxin-like X (SElX) is a unique superantigen with functional features of two major families of staphylococcal virulence factors.
PLoS Pathog., 13, 2017
|
|
1AH2
 
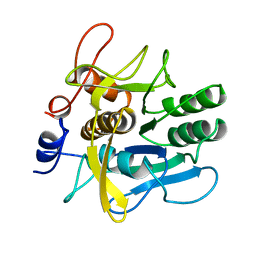 | | SERINE PROTEASE PB92 FROM BACILLUS ALCALOPHILUS, NMR, 18 STRUCTURES | | Descriptor: | SERINE PROTEASE PB92 | | Authors: | Boelens, R, Schipper, D, Martin, J.R, Karimi-Nejad, Y, Mulder, F, Zwan, J.V.D, Mariani, M. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of serine protease PB92 from Bacillus alcalophilus presents a rigid fold with a flexible substrate-binding site.
Structure, 5, 1997
|
|
4PCF
 
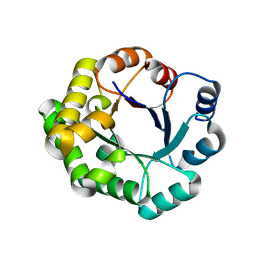 | |
4PCT
 
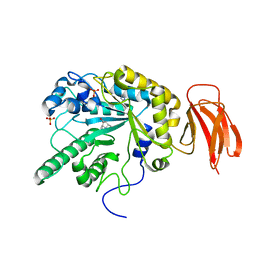 | | Crystal structure of a bacterial fucosidase with iminocyclitol (2S,3S,4R,5S)-3,4-dihydroxy-2-ethynyl-5-methylpyrrolidine | | Descriptor: | (2S,3S,4R,5S)-2-ethynyl-5-methylpyrrolidine-3,4-diol, Alpha-L-fucosidase, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2014-04-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploiting the Hydrophobic Terrain in Fucosidases with Aryl-Substituted Pyrrolidine Iminosugars.
Chembiochem, 16, 2015
|
|
4PRW
 
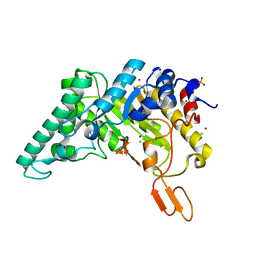 | | Xylanase T6 (XT6) from Geobacillus Stearothermophilus in complex with xylohexaose | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, SULFATE ION, ... | | Authors: | Dann, R.D, Solomon, H.V, Lansky, S, Ben-David, A, Lavid, N, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2014-03-06 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Xylanase T6 (XT6) from Geobacillus Stearothermophilus in complex with xylohexaose
To be Published
|
|
4PSC
 
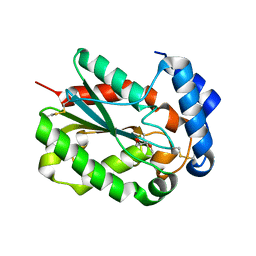 | | Structure of cutinase from Trichoderma reesei in its native form. | | Descriptor: | Carbohydrate esterase family 5, GLYCEROL | | Authors: | Roussel, A, Amara, S, Nyyssola, A, Mateos-Diaz, E, Blangy, S, Kontkanen, H, Westerholm-Parvinen, A, Carriere, F, Cambillau, C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Cutinase from Trichoderma reesei with a Lid-Covered Active Site and Kinetic Properties of True Lipases.
J.Mol.Biol., 426, 2014
|
|
4PSQ
 
 | | Crystal Structure of Retinol-Binding Protein 4 (RBP4) in complex with a non-retinoid ligand | | Descriptor: | (1-benzyl-1H-imidazol-4-yl)[4-(2-chlorophenyl)piperazin-1-yl]methanone, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Wang, Z, Johnstone, S, Walker, N. | | Deposit date: | 2014-03-07 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-assisted discovery of the first non-retinoid ligands for Retinol-Binding Protein 4.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4PT1
 
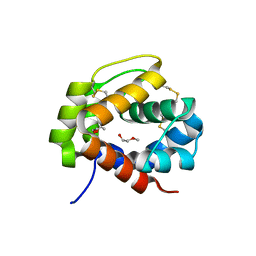 | |
6MRL
 
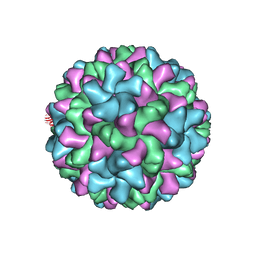 | | Cucumber Leaf Spot Virus | | Descriptor: | CALCIUM ION, p41 | | Authors: | Sherman, M.B, Smith, T.J. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes.
J.Virol., 94, 2020
|
|
4PG2
 
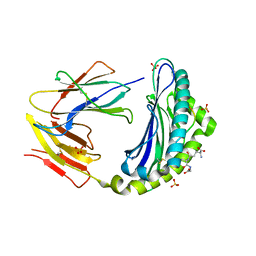 | |
1BH0
 
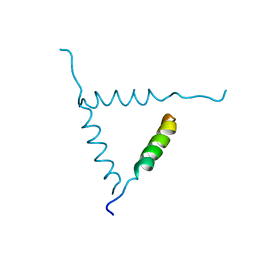 | | STRUCTURE OF A GLUCAGON ANALOG | | Descriptor: | GLUCAGON | | Authors: | Sturm, N.S, Lin, Y, Burley, S.K, Krstenansky, J.L, Ahn, J.-M, Azizeh, B.Y, Trivedi, D, Hruby, V.J. | | Deposit date: | 1998-06-11 | | Release date: | 1998-11-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies on positions 17, 18, and 21 replacement analogues of glucagon: the importance of charged residues and salt bridges in glucagon biological activity.
J.Med.Chem., 41, 1998
|
|
4P4J
 
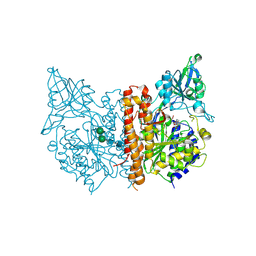 | |
4PG8
 
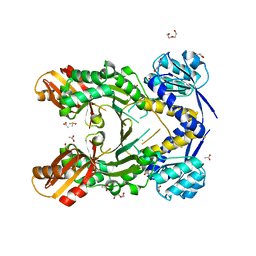 | |
6N3O
 
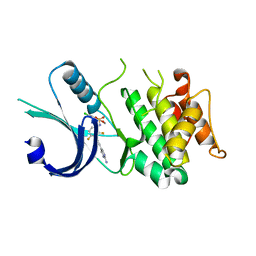 | | Identification of novel, potent and selective GCN2 inhibitors as first-in-class anti-tumor agents | | Descriptor: | N-{3-[(2-aminopyrimidin-5-yl)ethynyl]-2,4-difluorophenyl}-5-chloro-2-methoxypyridine-3-sulfonamide, eIF-2-alpha kinase GCN2 | | Authors: | Hoffman, I.D, Fujimoto, J, Kurasawa, O, Takagi, T, Klein, M.G, Kefala, G, Ding, S.C, Cary, D.R, Mizojiri, R. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Novel, Potent, and Orally Available GCN2 Inhibitors with Type I Half Binding Mode.
Acs Med.Chem.Lett., 10, 2019
|
|
4PHC
 
 | | Crystal Structure of a human cytosolic histidyl-tRNA synthetase, histidine-bound | | Descriptor: | GLYCEROL, HISTIDINE, Histidine--tRNA ligase, ... | | Authors: | Koh, C.Y, Wetzel, A.B, de van der Schueren, W.J, Hol, W.G.J. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Comparison of histidine recognition in human and trypanosomatid histidyl-tRNA synthetases.
Biochimie, 106, 2014
|
|
4P58
 
 | | Crystal structure of mouse comt bound to an inhibitor | | Descriptor: | 1',3'-dimethyl-1H,1'H-3,4'-bipyrazole, Catechol O-methyltransferase | | Authors: | Lanier, M. | | Deposit date: | 2014-03-15 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A fragment-based approach to identifying S-adenosyl-l-methionine -competitive inhibitors of catechol O-methyl transferase (COMT).
J.Med.Chem., 57, 2014
|
|
4PID
 
 | | Crystal structure of human adenovirus 2 protease with a weak pyrimidine nitrile inhibitor | | Descriptor: | ACETATE ION, N-benzyl-2-[(Z)-iminomethyl]pyrimidine-5-carboxamide, Pre-protein VI, ... | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
