5GGD
 
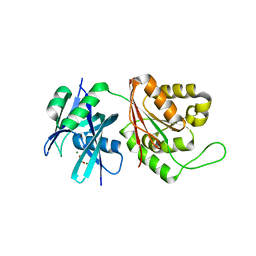 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with phosphate and magnesium ions (excess magnesium, II) | | Descriptor: | Hydrolase, NUDIX family protein, MAGNESIUM ION, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1VEQ
 
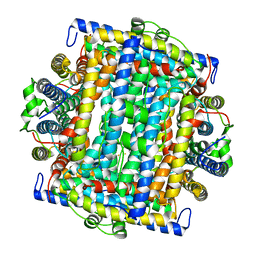 | | Mycobacterium smegmatis Dps Hexagonal form | | Descriptor: | FE (III) ION, starvation-induced DNA protecting protein | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-04-03 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules
J.Mol.Biol., 339, 2004
|
|
8PW3
 
 | |
7WDA
 
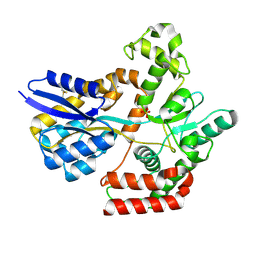 | | Crystal structure LpqY in complex with Trehalose from Mycobacterium tuberculosis | | Descriptor: | SULFATE ION, Trehalose-binding lipoprotein LpqY, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Sharma, D, Das, U. | | Deposit date: | 2021-12-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural analysis of LpqY, a substrate-binding protein from the SugABC transporter of Mycobacterium tuberculosis, provides insights into its trehalose specificity.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WCJ
 
 | | Crystal structure LpqY from Mycobacterium tuberculosis | | Descriptor: | SULFATE ION, Trehalose-binding lipoprotein LpqY | | Authors: | Sharma, D, Das, U. | | Deposit date: | 2021-12-20 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural analysis of LpqY, a substrate-binding protein from the SugABC transporter of Mycobacterium tuberculosis, provides insights into its trehalose specificity.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YY1
 
 | |
6Y8O
 
 | | Mycobacterium smegmatis GyrB 22kDa ATPase sub-domain in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
1VEL
 
 | | Mycobacterium smegmatis Dps tetragonal form | | Descriptor: | CADMIUM ION, SODIUM ION, SULFATE ION, ... | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-04-01 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules
J.Mol.Biol., 339, 2004
|
|
3K1D
 
 | | Crystal structure of glycogen branching enzyme synonym: 1,4-alpha-D-glucan:1,4-alpha-D-GLUCAN 6-glucosyl-transferase from mycobacterium tuberculosis H37RV | | Descriptor: | 1,4-alpha-glucan-branching enzyme | | Authors: | Pal, K, Kumar, S, Swaminathan, K. | | Deposit date: | 2009-09-27 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of full-length Mycobacterium tuberculosis H37Rv glycogen branching enzyme: insights of N-terminal beta-sandwich in substrate specificity and enzymatic activity
J.Biol.Chem., 285, 2010
|
|
1MQ7
 
 | | CRYSTAL STRUCTURE OF DUTPASE FROM MYCOBACTERIUM TUBERCULOSIS (RV2697C) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Sawaya, M.R, Chan, S, Segelke, B.W, Lekin, T, Heike, K, Cho, U.S, Naranjo, C, Perry, L.J, Yeates, T.O, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
6Y8L
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
1VEI
 
 | | Mycobacterium smegmatis Dps | | Descriptor: | FE (III) ION, SULFATE ION, starvation-induced DNA protecting protein | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray Analysis of Mycobacterium smegmatis Dps and a Comparative Study Involving Other Dps and Dps-like Molecules
J.Mol.Biol., 339, 2004
|
|
2A8X
 
 | | Crystal Structure of Lipoamide Dehydrogenase from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rajashankar, K.R, Bryk, R, Kniewel, R, Buglino, J.A, Nathan, C.F, Lima, C.D. | | Deposit date: | 2005-07-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and functional analysis of lipoamide dehydrogenase from Mycobacterium tuberculosis
J.Biol.Chem., 280, 2005
|
|
7BVB
 
 | |
7BVA
 
 | |
7YST
 
 | |
5GG6
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP | | Descriptor: | 1,2-ETHANEDIOL, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GGB
 
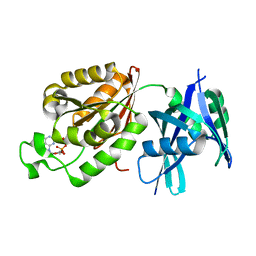 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGDP | | Descriptor: | 2'-deoxy-8-oxoguanosine 5'-(trihydrogen diphosphate), Hydrolase, NUDIX family protein | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3I4O
 
 | |
2QC3
 
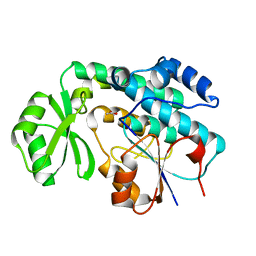 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
6S1T
 
 | |
6S2B
 
 | | Structure of beta-fructofuranosidase from Schwanniomyces occidentalis complexed with fructosyl-erythritol | | Descriptor: | (2~{S},3~{R})-4-[(2~{R},3~{S},4~{S},5~{R})-2,5-bis(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxybutane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-06-20 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | New insights into the molecular mechanism behind mannitol and erythritol fructosylation by beta-fructofuranosidase from Schwanniomyces occidentalis.
Sci Rep, 11, 2021
|
|
2O2J
 
 | |
2G1K
 
 | | Crystal structure of Mycobacterium tuberculosis shikimate kinase in complex with shikimate at 1.75 angstrom resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Gan, J, Gu, Y, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2006-02-14 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Shikimate Kinase in Complex with Shikimic Acid and an ATP Analogue.
Biochemistry, 45, 2006
|
|
2AQH
 
 | | Crystal structure of Isoniazid-resistant I21V Enoyl-ACP(CoA) reductase mutant enzyme from Mycobacterium tuberculosis in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Oliveira, J.S, Pereira, J.H, Rodrigues, N.C, Canduri, F, de Souza, O.N, Basso, L.A, de Azevedo Jr, W.F, Santos, D.S. | | Deposit date: | 2005-08-18 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystallographic and Pre-steady-state Kinetics Studies on Binding of NADH to Wild-type and Isoniazid-resistant Enoyl-ACP(CoA) Reductase Enzymes from Mycobacterium tuberculosis.
J.Mol.Biol., 359, 2006
|
|
