5A2Z
 
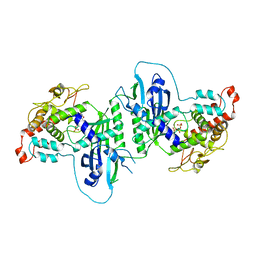 | | Crystal structure of mtPAP in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
8YRC
 
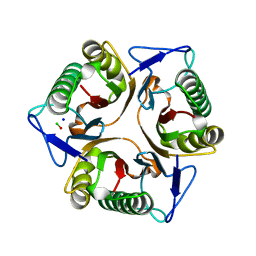 | | Chlorinated YabJ from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, OXYGEN ATOM, SODIUM ION, ... | | Authors: | Jeong, C, Kim, H.J. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | YabJ from Staphylococcus aureus entraps chlorides within its pocket.
Biochem.Biophys.Res.Commun., 710, 2024
|
|
8CQB
 
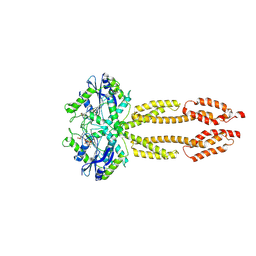 | |
5A2X
 
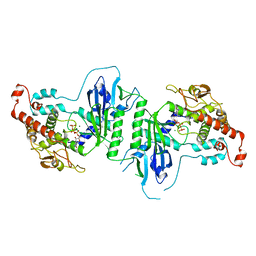 | | Crystal structure of mtPAP in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
2C52
 
 | | Structural diversity in CBP p160 complexes | | Descriptor: | CREB-BINDING PROTEIN, NUCLEAR RECEPTOR COACTIVATOR 1 | | Authors: | Waters, L.C, Yue, B, Veverka, V, Renshaw, P.S, Bramham, J, Matsuda, S, Frenkiel, T, Kelly, G, Muskett, F.W, Carr, M.D, Heery, D.M. | | Deposit date: | 2005-10-25 | | Release date: | 2006-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in p160/CREB-binding protein coactivator complexes.
J. Biol. Chem., 281, 2006
|
|
8YL9
 
 | | Crystal structures of terpene synthases complexed with a substrate mimic | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE, ... | | Authors: | Xu, M, Ma, M. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights Into the Terpene Cyclization Domains of Two Fungal Sesterterpene Synthases and Enzymatic Engineering for Sesterterpene Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1ESU
 
 | | S235A MUTANT OF TEM1 BETA-LACTAMASE | | Descriptor: | BETA-LACTAMASE, SULFATE ION | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-11 | | Release date: | 2000-05-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TEM1 beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1J54
 
 | | Structure of the N-terminal exonuclease domain of the epsilon subunit of E.coli DNA polymerase III at pH 5.8 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
6CON
 
 | | Crystal structure of Mycobacterium tuberculosis IpdAB | | Descriptor: | CoA-transferase subunit alpha, CoA-transferase subunit beta | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CPZ
 
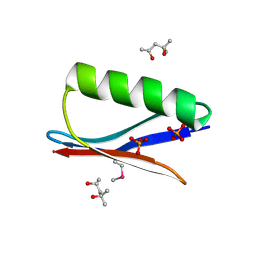 | | Selenomethionine mutant (I6Sem) of protein GB1 examined by X-ray diffraction | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Immunoglobulin G-binding protein G, ... | | Authors: | Chen, Q, Rozovsky, S. | | Deposit date: | 2018-03-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | 77Se NMR Probes the Protein Environment of Selenomethionine.
J.Phys.Chem.B, 124, 2020
|
|
1ENK
 
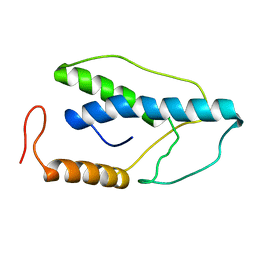 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
8XLM
 
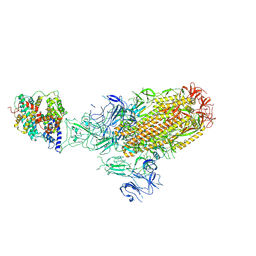 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-12-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
6CSM
 
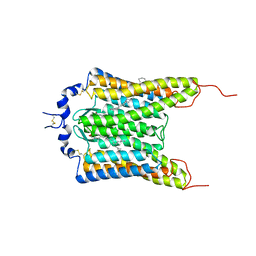 | | Crystal structure of the natural light-gated anion channel GtACR1 | | Descriptor: | GtACR1, OLEIC ACID, RETINAL | | Authors: | Kato, H.E, Kim, Y, Yamashita, K, Kobilka, B.K, Deisseroth, K. | | Deposit date: | 2018-03-21 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanisms of selectivity and gating in anion channelrhodopsins.
Nature, 561, 2018
|
|
5WD6
 
 | | bovine salivary protein form 30b | | Descriptor: | CALCIUM ION, Short palate, lung and nasal epithelium carcinoma-associated protein 2B | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
2BLX
 
 | | HEWL before a high dose x-ray "burn" | | Descriptor: | LYSOZYME C, TETRAETHYLENE GLYCOL | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving Radiation-Damage Substructures for Rip.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ENI
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
6CUH
 
 | | Crystal structure of the unliganded BC8B TCR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shahine, A.E, Rossjohn, J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids.
Nat Commun, 10, 2019
|
|
7EV8
 
 | |
6CV7
 
 | |
1EOS
 
 | | CRYSTAL STRUCTURE OF RIBONUCLEASE A COMPLEXED WITH URIDYLYL(2',5')GUANOSINE (PRODUCTIVE BINDING) | | Descriptor: | RIBONUCLEASE PANCREATIC, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | Authors: | Vitagliano, L, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 2000-03-24 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Productive and nonproductive binding to ribonuclease A: X-ray structure of two complexes with uridylyl(2',5')guanosine.
Protein Sci., 9, 2000
|
|
8XLN
 
 | | Structure of the SARS-CoV-2 EG.5.1 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-12-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
2BN2
 
 | |
1ESQ
 
 | | CRYSTAL STRUCTURE OF THIAZOLE KINASE MUTANT (C198S) WITH ATP AND THIAZOLE PHOSPHATE. | | Descriptor: | 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, HYDROXYETHYLTHIAZOLE KINASE, ... | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-04-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
6CXB
 
 | |
2BVE
 
 | | Structure of the N-terminal of Sialoadhesin in complex with 2-Phenyl- Prop5Ac | | Descriptor: | SIALOADHESIN, benzyl 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosidonic acid | | Authors: | Zaccai, N.R, May, A.P, Robinson, R.C, Burtnick, L.D, Crocker, P, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2005-06-27 | | Release date: | 2006-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and in Silico Analysis of the Sialoside-Binding Characteristics of the Siglec Sialoadhesin.
J.Mol.Biol., 365, 2007
|
|
