8A66
 
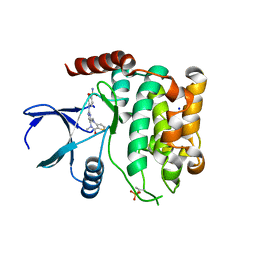 | | Crystal structure of MST2 in complex with XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, SODIUM ION, Serine/threonine-protein kinase 3 36kDa subunit | | Authors: | Nawrotek, A, Vuillard, L, Miallau, L, Weber, C. | | Deposit date: | 2022-06-16 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of the Kelch domain of human Keap1in complex with ligand S217879
To Be Published
|
|
7ZKJ
 
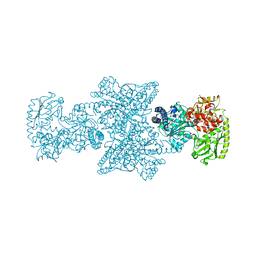 | | CODH/ACS complex of C. hydrogenoformans | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Ruickoldt, J, Jeoung, J.-H, Basak, Y, Domnik, L, Dobbek, H. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | On the Kinetics of CO2 Reduction by Ni, Fe-CO Dehydrogenases
ACS Catal., 2022
|
|
7ZHE
 
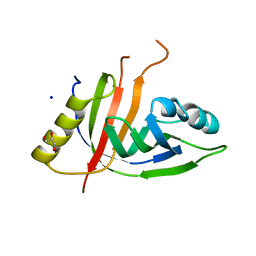 | | Crystal structure of CtaZ from Ruminiclostridium cellulolyticum | | Descriptor: | GLYCEROL, SODIUM ION, Transcription activator effector binding | | Authors: | Gude, F, Molloy, E.M, Horch, T, Dell, M, Dunbar, K.L, Krabbe, J, Groll, M, Hertweck, C. | | Deposit date: | 2022-04-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Specialized Polythioamide-Binding Protein Confers Antibiotic Self-Resistance in Anaerobic Bacteria.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5WKE
 
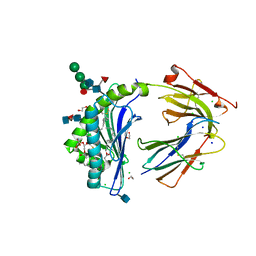 | | Crystal Structure of Human CD1b in Complex with PS | | Descriptor: | (19S,22S,25R)-25-amino-22-hydroxy-16,22,26-trioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shahine, A, Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A molecular basis of human T cell receptor autoreactivity toward self-phospholipids.
Sci Immunol, 2, 2017
|
|
5WPT
 
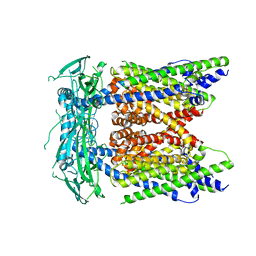 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed II conformation at 3.75 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
7L6C
 
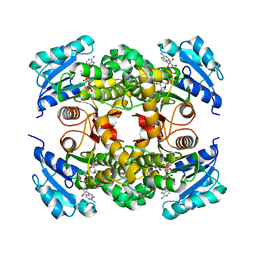 | |
7KUQ
 
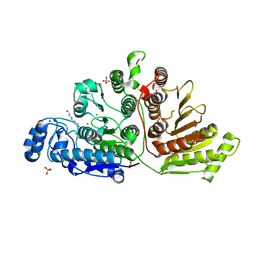 | |
4LSS
 
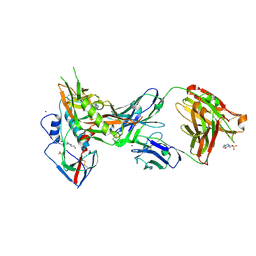 | | Crystal structure of broadly and potently neutralizing antibody VRC01 in complex with HIV-1 clade A strain KER_2018_11 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEAVY CHAIN OF ANTIBODY VRC01, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
7L35
 
 | | Human DNA Ligase 1 - R771W nicked DNA complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tumbale, P.P, Williams, R.S, Schellenberg, M.S. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | LIG1 syndrome mutations remodel a cooperative network of ligand binding interactions to compromise ligation efficiency.
Nucleic Acids Res., 49, 2021
|
|
1IP2
 
 | | G48A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
4LZ1
 
 | | X-ray structure of the complex between human thrombin and the TBA deletion mutant lacking thymine 12 nucleobase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, ... | | Authors: | Pica, A, Russo Krauss, I, Merlino, A, Sica, F. | | Deposit date: | 2013-07-31 | | Release date: | 2014-01-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer.
Febs J., 280, 2013
|
|
1IX0
 
 | | I59A-3SS human lysozyme | | Descriptor: | SODIUM ION, lysozyme | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2002-06-06 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Buried water molecules contribute to the conformational stability of a protein
PROTEIN ENG., 16, 2003
|
|
4M4V
 
 | |
7KGP
 
 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N316-324 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KUR
 
 | |
7KUT
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 10 H137A Mutant in Complex with N-Acetylputrescine (Tetrahedral Intermediate) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-aminobutyl)amino]ethane-1,1-diol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-11-25 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystallographic Snapshots of Substrate Binding in the Active Site of Histone Deacetylase 10.
Biochemistry, 60, 2021
|
|
7KIA
 
 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 19 | | Descriptor: | 1-[4-(4-{4-(4-methylpiperazin-1-yl)-6-[(3-methyl-1H-pyrazol-5-yl)amino]pyrimidin-2-yl}phenyl)piperidin-1-yl]prop-2-en-1-one, CITRATE ANION, Fibroblast growth factor receptor 2, ... | | Authors: | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
7KV2
 
 | | Surface glycan-binding protein A from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RagB/SusD family nutrient uptake outer membrane protein, ... | | Authors: | Tamura, K, Brumer, H, Van Petegem, F. | | Deposit date: | 2020-11-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct protein architectures mediate species-specific beta-glucan binding and metabolism in the human gut microbiota.
J.Biol.Chem., 296, 2021
|
|
4HMD
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7KZA
 
 | |
2D4D
 
 | | The Crystal Structure of human beta2-microglobulin, L39W W60F W95F Mutant | | Descriptor: | Beta-2-microglobulin, SODIUM ION | | Authors: | Iwata, K, Matsuura, T, Nakagawa, A, Goto, Y. | | Deposit date: | 2005-10-17 | | Release date: | 2006-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformation of Amyloid Fibrils of beta2-Microglobulin Probed by Tryptophan Mutagenesis
J.Biol.Chem., 281, 2006
|
|
4LOR
 
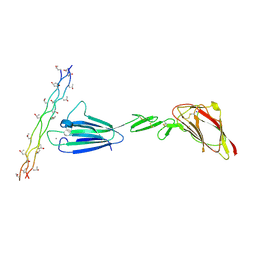 | | C1s CUB1-EGF-CUB2 in complex with a collagen-like peptide from C1q | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1s subcomponent heavy chain, ... | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HME
 
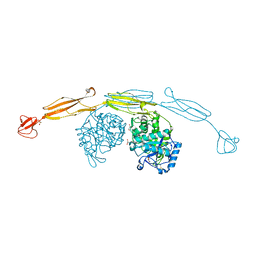 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HQO
 
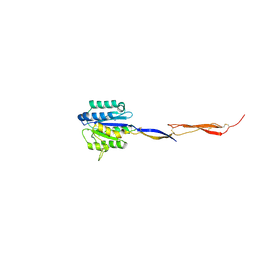 | | Crystal structure of Plasmodium vivax TRAP protein | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7L1A
 
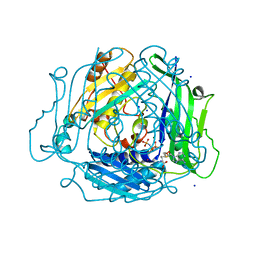 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine and inhibitor, di-imido triphosphate (PNPNP) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, MAGNESIUM ION, ... | | Authors: | Niland, C.N, Fedorov, E, Schramm, V.L, Ghosh, A. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism and Inhibition of Human Methionine Adenosyltransferase 2A.
Biochemistry, 60, 2021
|
|
