6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
8CB4
 
 | |
8CBG
 
 | |
8CB5
 
 | |
8CB8
 
 | |
8CBP
 
 | |
8CB7
 
 | |
8CBI
 
 | |
6ZOP
 
 | | Structure of the cysteine-rich domain of PiggyMac, a domesticated PiggyBac transposase involved in programmed genome rearrangements | | Descriptor: | DDE_Tnp_1_7 domain-containing protein, ZINC ION | | Authors: | Bessa, L, Guerineau, M, Moriau, S, Lescop, E, Bontems, F, Mathy, N, Guittet, E, Bischerour, J, Betermier, M, Morellet, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | SOLUTION NMR | | Cite: | The unusual structure of the PiggyMac cysteine-rich domain reveals zinc finger diversity in PiggyBac-related transposases.
Mob DNA, 12, 2021
|
|
4WU9
 
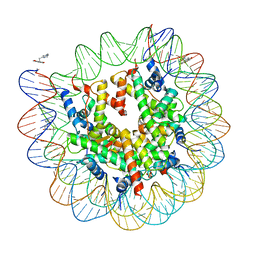 | | Structure of cisPtNAP-NCP145 | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Chua, E.Y.D, Davey, G.E, Chin, C.F, Droge, P, Ang, W.H, Davey, C.A. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stereochemical control of nucleosome targeting by platinum-intercalator antitumor agents.
Nucleic Acids Res., 43, 2015
|
|
8DAF
 
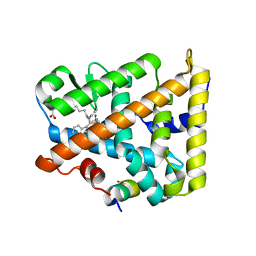 | | Human SF-1 LBD bound to synthetic agonist 6N-10CA and bacterial phospholipid | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, ... | | Authors: | D'Agostino, E.H, Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparison of activity, structure, and dynamics of SF-1 and LRH-1 complexed with small molecule modulators.
J.Biol.Chem., 299, 2023
|
|
7PFM
 
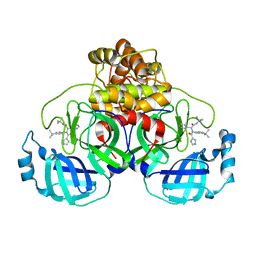 | | A SARS-CoV2 major protease non-covalent ligand structure determined to 2.0 A resolution | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-N-(4-tert-butylphenyl)-1H-imidazole-5-carboxamide, Replicase polyprotein 1ab | | Authors: | Moche, M, Moodie, L, Strandback, E, Nyman, T, Sandstrom, A, Akaberi, D, Lennerstrand, J. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
6VCJ
 
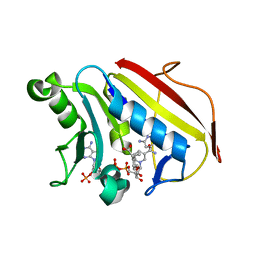 | | Crystal structure of hsDHFR in complex with NADP+, DAP, and R-naproxen | | Descriptor: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Pedersen, L.C, London, R.E, Gabel, S.A, Krahn, J.M, DeRose, E.F. | | Deposit date: | 2019-12-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The Structural Basis for Nonsteroidal Anti-Inflammatory Drug Inhibition of Human Dihydrofolate Reductase.
J.Med.Chem., 63, 2020
|
|
7A4S
 
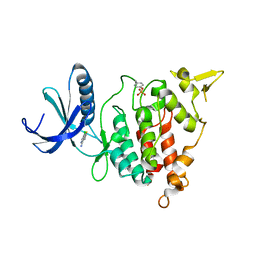 | | Structure of DYRK1A in complex with compound 2 | | Descriptor: | 7-chlorothieno[3,2-c]pyridin-4-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
7A4Z
 
 | | Structure of DYRK1A in complex with compound 4 | | Descriptor: | 3-(4-methoxyphenyl)-1~{H}-pyrazol-5-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
1JLB
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
6GO1
 
 | | Crystal Structure of a Bacillus anthracis peptidoglycan deacetylase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Polysaccharide deacetylase-like protein, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2018-06-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The putative polysaccharide deacetylase Ba0331: cloning, expression, crystallization and structure determination.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6ORQ
 
 | | Modified BG505 SOSIP-based immunogen RC1 in complex with the elicited V3-glycan patch antibody Ab275MUR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab275MUR antibody Fab heavy chain, ... | | Authors: | Abernathy, M.E, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques.
Nature, 570, 2019
|
|
4TNW
 
 | | C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab and POPC in a lipid-modulated conformation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avermectin-sensitive glutamate-gated chloride channel GluCl alpha, ... | | Authors: | Althoff, T, Hibbs, R.E, Banerjee, S, Gouaux, E. | | Deposit date: | 2014-06-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors.
Nature, 512, 2014
|
|
1OH5
 
 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A C:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*CP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
7PKA
 
 | | Synechocystis sp. PCC6803 glutathione transferase Chi 1, GSOH bound | | Descriptor: | Glutathione S-transferase, POTASSIUM ION, S-Hydroxy-Glutathione | | Authors: | Didierjean, C, Mocchetti, E, Hecker, A, Favier, F. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure and functions of glutathione transferase Chi 1 from cyanobacterium Synechocystis sp. PCC 6803
To Be Published
|
|
7TTJ
 
 | | Stable-5-LOX elongated Ha2 | | Descriptor: | Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Gilbert, N.C, Newcomer, M.E. | | Deposit date: | 2022-02-01 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Helical remodeling augments 5-lipoxygenase activity in the synthesis of proinflammatory mediators.
J.Biol.Chem., 298, 2022
|
|
1OH6
 
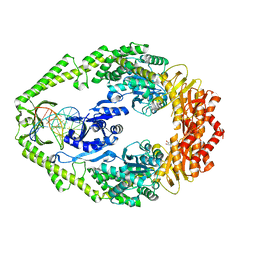 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN A:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*AP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP* CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP* CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
7PHZ
 
 | | Crystal structure of X77 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P2(1)2(1)2(1). | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
7U67
 
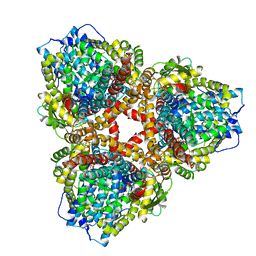 | | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and GTP | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, GUANOSINE-5'-TRIPHOSPHATE, Inhibitor of dGTPase, ... | | Authors: | Klemm, B.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism by which T7 bacteriophage protein Gp1.2 inhibits Escherichia coli dGTPase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
