4IXX
 
 | | Crystal structure of S213G variant DAH7PS without Tyr bound from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, MANGANESE (II) ION, SULFATE ION | | Authors: | Cross, P.J, Pietersma, A.L, Allison, T.M, Wilson-Coutts, S.M, Cochrane, F.C, Parker, E.J. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.
Protein Sci., 22, 2013
|
|
3OUP
 
 | |
3F0B
 
 | | Staphylococcus aureus dihydrofolate reductase complexed with NADPH and 2,4-Diamino-5-[3-(3-methoxy-5-phenylphenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3R)-3-(5-methoxybiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2008-10-24 | | Release date: | 2009-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance.
J.Mol.Biol., 387, 2009
|
|
3F4R
 
 | | Crystal structure of Wolbachia pipientis alpha-DsbA1 | | Descriptor: | PENTAETHYLENE GLYCOL, Putative uncharacterized protein, TRIETHYLENE GLYCOL | | Authors: | Kurz, M, Heras, B, Martin, J.L. | | Deposit date: | 2008-11-02 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of the Oxidoreductase alpha-DsbA1 from Wolbachia pipientis
ANTIOXID.REDOX SIGNAL., 11, 2009
|
|
3F0E
 
 | |
3F1K
 
 | |
3OYS
 
 | | Human Carbonic Anhydrase II complexed with 2-{[4-AMINO-3-(3-HYDROXYPROP-1-YN-1-YL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-1-YL]METHYL}-5-METHYL-3-(2-METHYLPHENYL)QUINAZOLIN-4(3H)-ONE | | Descriptor: | 2-phenyl-N-(4-sulfamoylphenyl)acetamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Aggarwal, M, McKenna, R. | | Deposit date: | 2010-09-23 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Anticonvulsant 4-aminobenzenesulfonamide derivatives with branched-alkylamide moieties: X-ray crystallography and inhibition studies of human carbonic anhydrase isoforms I, II, VII, and XIV.
J.Med.Chem., 54, 2011
|
|
4LBS
 
 | | Crystal structure of human AR complexed with NADP+ and {2-[(4-bromo-2,6-difluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-bromo-2,6-difluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.76 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
4J1O
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound l-proline betaine (substrate) | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, GLYCEROL, IODIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-01 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
4J2C
 
 | | GARP-SNARE Interaction | | Descriptor: | Syntaxin-6, Vacuolar protein sorting-associated protein 51 homolog | | Authors: | Abascal-Palacios, G, Schindler, C, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the interaction of the Golgi-Associated Retrograde Protein Complex with the t-SNARE Syntaxin 6.
Structure, 21, 2013
|
|
4LFL
 
 | |
4LFS
 
 | | High resolution x-ray structure of racemic ShK toxin | | Descriptor: | CHLORIDE ION, GLYCEROL, Potassium channel toxin ShK | | Authors: | Dang, B, Kubota, T, Mandal, K, Bezanilla, F, Kent, S.B.H. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Native Chemical Ligation at Asx-Cys, Glx-Cys: Chemical Synthesis and High-Resolution X-ray Structure of ShK Toxin by Racemic Protein Crystallography.
J.Am.Chem.Soc., 135, 2013
|
|
4M47
 
 | | structure of human DNA polymerase complexed with 8-BrG in the template base paired with incoming non-hydrolyzable GTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-06 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4J3S
 
 | | Crystal structure of barley limit dextrinase soaked with 300mM maltotetraose | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Sim, L, Windahl, M.S, Moeller, M.S, Henriksen, A. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase
J.Mol.Biol., 427, 2015
|
|
4J3V
 
 | | Crystal structure of barley limit dextrinase in complex with a branched thio-linked hexasaccharide | | Descriptor: | CALCIUM ION, CHLORIDE ION, IODIDE ION, ... | | Authors: | Sim, L, Windahl, M.S, Moeller, M.S, Henriksen, A. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase
J.Mol.Biol., 427, 2015
|
|
3P2F
 
 | | Crystal structure of TofI in an apo form | | Descriptor: | AHL synthase | | Authors: | Yu, S, Rhee, S. | | Deposit date: | 2010-10-02 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibitor binding to an N-acyl-homoserine lactone synthase
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4IRN
 
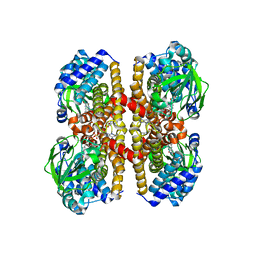 | | Crystal Structure of the Prolyl Acyl Carrier Protein Oxidase AnaB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Prolyl-ACP dehydrogenase | | Authors: | Moncoq, K, Mann, S, Regad, L, Mejean, A, Ploux, O. | | Deposit date: | 2013-01-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the prolyl-acyl carrier protein oxidase involved in the biosynthesis of the cyanotoxin anatoxin-a.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4ISU
 
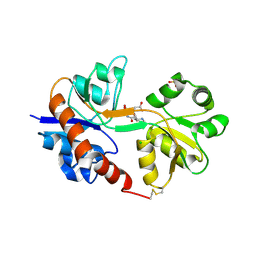 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (2R)-IKM-159 at 2.3A resolution. | | Descriptor: | (4aS,5aR,6R,8aS,8bS)-5a-(carboxymethyl)-8-oxo-2,4a,5a,6,7,8,8a,8b-octahydro-1H-pyrrolo[3',4':4,5]furo[3,2-b]pyridine-6-carboxylic acid, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Juknaite, L, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Studies on an (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA) receptor antagonist IKM-159: asymmetric synthesis, neuroactivity, and structural characterization.
J.Med.Chem., 56, 2013
|
|
4IT5
 
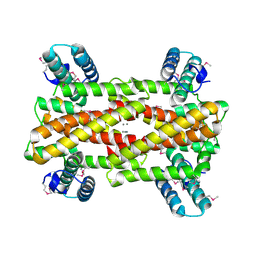 | | Chaperone HscB from Vibrio cholerae | | Descriptor: | CALCIUM ION, Co-chaperone protein HscB homolog | | Authors: | Osipiuk, J, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-17 | | Release date: | 2013-01-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Chaperone HscB from Vibrio cholerae.
To be Published
|
|
4ITM
 
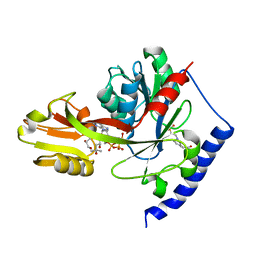 | | Crystal structure of "apo" form LpxK from Aquifex aeolicus in complex with ATP at 2.2 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1994 Å) | | Cite: | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
4MC1
 
 | | HIV protease in complex with SA526P | | Descriptor: | (3S)-tetrahydrofuran-3-yl {(2S,3R)-4-[(4S)-4-tert-butyl-7-fluoro-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Ganguly, A.K, Alluri, S.S, Wang, C, Antropow, A, White, A, Caroccia, D, Biswas, D, Kang, E, Zhang, L, Carroll, S.S, Burlein, C, Munshi, V, Orth, P, Strickland, C. | | Deposit date: | 2013-08-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Optimization of Cyclic Sulfonamide based Novel HIV-1 Protease Inhibitors to Pico Molar Affinities guided by X-ray Crystallographic Analysis
Tetrahedron, 2014
|
|
3P5N
 
 | |
4IV5
 
 | |
3P5V
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3FFU
 
 | |
