6W43
 
 | | APE1 AP-endonuclease product complex R237C | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
2V79
 
 | | Crystal Structure of the N-terminal domain of DnaD from Bacillus Subtilis | | Descriptor: | CHLORIDE ION, DNA REPLICATION PROTEIN DNAD, SODIUM ION | | Authors: | Schneider, S, Zhang, W, Soultanas, P, Paoli, M. | | Deposit date: | 2007-07-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the N-Terminal Oligomerization Domain of Dnad Reveals a Unique Tetramerization Motif and Provides Insights Into Scaffold Formation.
J.Mol.Biol., 376, 2008
|
|
8TFL
 
 | | Ricin in complex with Fab SylH3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, NONAETHYLENE GLYCOL, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Basis of Antibody-Mediated Inhibition of Ricin Toxin Attachment to Host Cells.
Biochemistry, 62, 2023
|
|
3LL9
 
 | |
8TFH
 
 | | Ricin in complex with Fab JB4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, JB4 monoclonal antibody heavy chain, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Basis of Antibody-Mediated Inhibition of Ricin Toxin Attachment to Host Cells.
Biochemistry, 62, 2023
|
|
4G1P
 
 | | Structural and Mechanistic Basis of Substrate Recognition by Novel Di-peptidase Dug1p From Saccharomyces cerevisiae | | Descriptor: | CYSTEINE, Cys-Gly metallodipeptidase DUG1, GLYCINE, ... | | Authors: | Singh, A.K, Singh, M, Pandya, V.K, Singh, V, Mittal, M, Kumaran, S. | | Deposit date: | 2012-07-11 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Structural and Mechanistic Basis of Substrate Recognition by Novel Di-peptidase Dug1p From Saccromyces cerevesiae
To be published
|
|
3L5W
 
 | | Crystal structure of the complex between IL-13 and C836 FAB | | Descriptor: | C836 HEAVY CHAIN, C836 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
6W4I
 
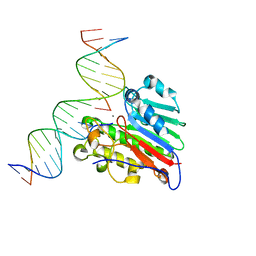 | | APE1 Y269A product complex with abasic DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop.
Nucleic Acids Res., 48, 2020
|
|
4E2F
 
 | |
3LHP
 
 | |
4DY9
 
 | |
4GIG
 
 | |
3L7E
 
 | | Crystal structure of ANTI-IL-13 antibody C836 | | Descriptor: | ACETATE ION, C836 HEAVY CHAIN, C836 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antigen recognition by antibody C836 through adjustment of VL/VH packing
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2J2I
 
 | | Crystal Structure of the humab PIM1 in complex with LY333531 | | Descriptor: | (9R)-9-[(DIMETHYLAMINO)METHYL]-6,7,10,11-TETRAHYDRO-9H,18H-5,21:12,17-DIMETHENODIBENZO[E,K]PYRROLO[3,4-H][1,4,13]OXADIA ZACYCLOHEXADECINE-18,20-DIONE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1, SULFATE ION | | Authors: | Debreczeni, J.E, Bullock, A.N, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Weigelt, J, Knapp, S. | | Deposit date: | 2006-08-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
3LQU
 
 | |
2VOE
 
 | | Crystal structure of Rv2780 from M. tuberculosis H37Rv | | Descriptor: | ALANINE DEHYDROGENASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Mycobacterium Tuberculosis Secretory Antigen Alanine Dehydrogenase (Rv2780) in Apo and Ternary Complex Forms Captures "Open" and "Closed" Enzyme Conformations.
Proteins, 72, 2008
|
|
3L7F
 
 | | Structure of IL-13 antibody H2L6, A humanized variant OF C836 | | Descriptor: | CALCIUM ION, H2L6 HEAVY CHAIN, H2L6 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3LH2
 
 | |
6VX2
 
 | |
3LUG
 
 | |
6W0U
 
 | |
8T52
 
 | |
6W42
 
 | |
6W11
 
 | |
6W6T
 
 | | WT HIV-1 Protease in Complex with Phosphonated UMass6 (PU6) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2020-03-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | To Be Determined
To Be Published
|
|
