5Q0C
 
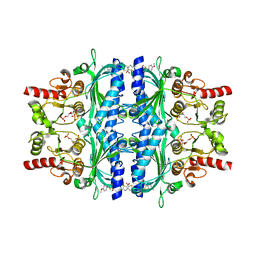 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(4-methoxyphenyl)thiophen-2-yl]sulfonylurea and with fructose-2,6-diphosphate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(4-methoxyphenyl)thiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(4-methoxyphenyl)thiophen-2-yl]sulfonylurea and with f2,6p
To be published
|
|
5Q01
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(4,5-dichloro-2-methylpyrazol-3-yl)oxyphenyl]sulfonylurea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-4-[(3,4-dichloro-1-methyl-1H-pyrazol-5-yl)oxy]benzene-1-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[4-(4,5-dichloro-2-methylpyrazol-3-yl)oxyphenyl]sulfonylurea
To be published
|
|
2GUY
 
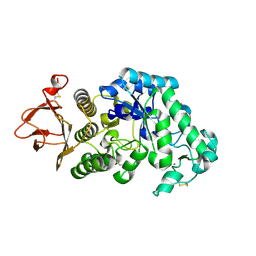 | |
2NY5
 
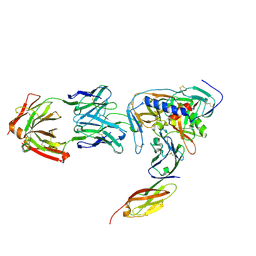 | | HIV-1 gp120 Envelope Glycoprotein (M95W, W96C, I109C, T257S, V275C, S334A, S375W, Q428C, A433M) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
2M1A
 
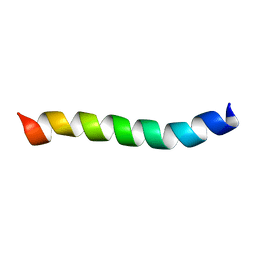 | | HIV-1 Rev ARM peptide (residues T34-R50) | | Descriptor: | HIV-1 Rev arginine-rich motif (ARM) | | Authors: | Casu, F, Duggan, B.M, Hennig, M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Arginine-Rich RNA-Binding Motif of HIV-1 Rev Is Intrinsically Disordered and Folds upon RRE Binding.
Biophys.J., 105, 2013
|
|
2NUI
 
 | | X-ray Structure of synthetic [D83A]RNase A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Boerema, D.J, Tereshko, V.A, Kent, S.B.H. | | Deposit date: | 2006-11-09 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Total synthesis by modern chemical ligation methods and high resolution (1.1 A) X-ray structure of ribonuclease A
Biopolymers, 90, 2008
|
|
2OJ9
 
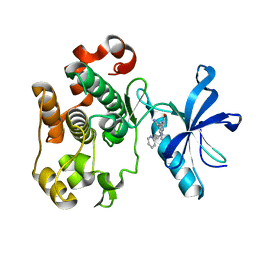 | | Structure of IGF-1R kinase domain complexed with a benzimidazole inhibitor | | Descriptor: | 3-[5-(1H-IMIDAZOL-1-YL)-7-METHYL-1H-BENZIMIDAZOL-2-YL]-4-[(PYRIDIN-2-YLMETHYL)AMINO]PYRIDIN-2(1H)-ONE, Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) | | Authors: | Sack, J.S, Jacobson, B.L. | | Deposit date: | 2007-01-12 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and initial SAR of 3-(1H-benzo[d]imidazol-2-yl)pyridin-2(1H)-ones as inhibitors of insulin-like growth factor 1-receptor (IGF-1R).
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2MWK
 
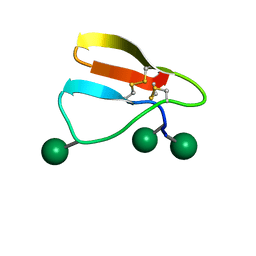 | | Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1, Ser3, and Ser14 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
2MWJ
 
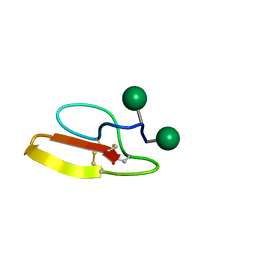 | | Solution structure of Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1 and Ser3 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
2NY1
 
 | | HIV-1 gp120 Envelope Glycoprotein (I109C, T257S, S334A, S375W, Q428C) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
2ONF
 
 | |
2OQM
 
 | |
2LAV
 
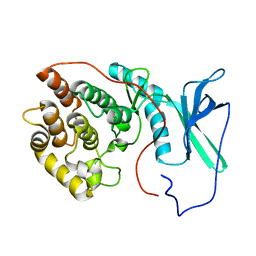 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
1C1B
 
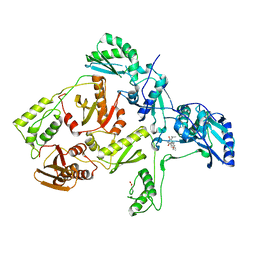 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GCA-186 | | Descriptor: | 6-(3',5'-DIMETHYLBENZYL)-1-ETHOXYMETHYL-5-ISOPROPYLURACIL, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Hopkins, A.L, Ren, J, Tanaka, H, Baba, B, Okamato, M, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-21 | | Release date: | 2000-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
1C0T
 
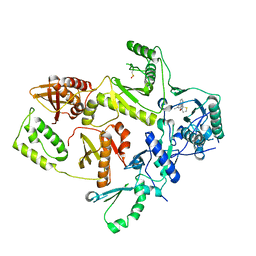 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+21.1326 | | Descriptor: | (R)-(+)9B-(3-METHYL)PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-5(9BH)-ONE, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
2MB3
 
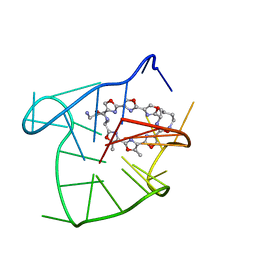 | | Solution structure of an intramolecular (3+1) human telomeric G-quadruplex bound to a telomestatin derivative | | Descriptor: | (12S,27S)-12,27-bis(4-aminobutyl)-4,30-dimethyl-3,7,14,18,22,29-hexaoxa-11,26,31,32,33,34,35,36-octaazaheptacyclo[26.2. 1.1~2,5~.1~6,9~.1~13,16~.1~17,20~.1~21,24~]hexatriaconta-1(30),2(36),4,6(35),8,13(34),15,17(33),19,21(32),23,28(31)-dode caene-10,25-dione, DNA_(5'-D(*TP*TP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*A)-3') | | Authors: | Chung, W.J, Heddi, B, Tera, M, Iida, K, Nagasawa, K, Phan, A.T. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular (3 + 1) human telomeric g-quadruplex bound to a telomestatin derivative.
J.Am.Chem.Soc., 135, 2013
|
|
2D0G
 
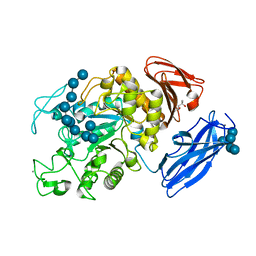 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P5, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2GVY
 
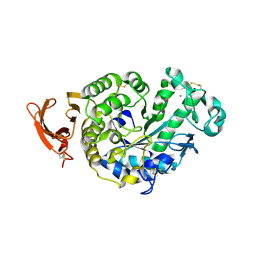 | |
2BGN
 
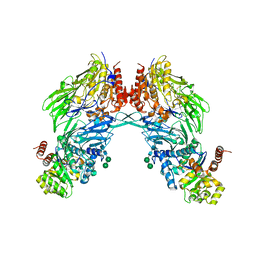 | | HIV-1 Tat protein derived N-terminal nonapeptide Trp2-Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2005-01-03 | | Release date: | 2005-01-27 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structures of HIV-1 Tat-Derived Nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) Bound to the Active Site of Dipeptidyl-Peptidase Iv (Cd26)
J.Biol.Chem., 280, 2005
|
|
2JIH
 
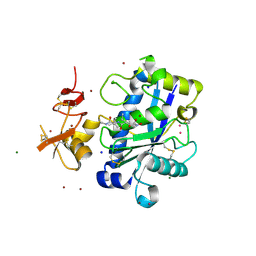 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (complex-form) | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAMTS-1, CADMIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains.
J. Mol. Biol., 373, 2007
|
|
1C1C
 
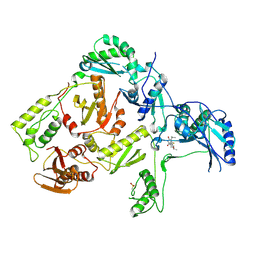 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-6123 | | Descriptor: | 6-(cyclohexylsulfanyl)-1-(ethoxymethyl)-5-(1-methylethyl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Hopkins, A.L, Ren, J, Tanaka, H, Baba, M, Okamato, M, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-21 | | Release date: | 2000-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
7K54
 
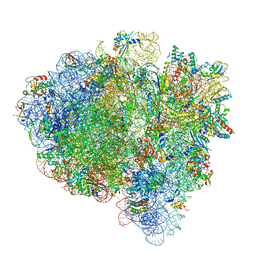 | | Mid-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure II-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
1RP8
 
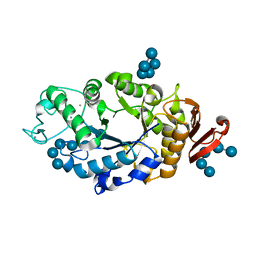 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) inactive mutant d180a in complex with maltoheptaose | | Descriptor: | Alpha-amylase type 1 isozyme, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-12-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
7K53
 
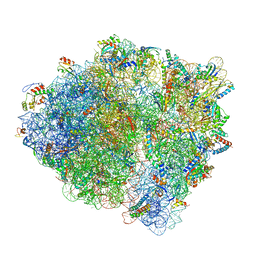 | | Pre-translocation +1-frameshifting(CCC-A) complex (Structure I-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K55
 
 | | Near post-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure III-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
