5ZBX
 
 | | The crystal structure of the nucleosome containing histone H3.1 CATD(V76Q, K77D) | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Takagi, H, Kurumizaka, H. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism.
Nat Commun, 10, 2019
|
|
7NEW
 
 | | Fucosylated heterochiral linear peptide Fdln69 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 2.0 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Personne, H, Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A mixed chirality alpha-helix in a stapled bicyclic and a linear antimicrobial peptide revealed by X-ray crystallography.
Rsc Chem Biol, 2, 2021
|
|
8XH1
 
 | |
8XH0
 
 | |
4PRE
 
 | | Crystal structure of a HLA-B*35:08-HPVG-Q5 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
5WNK
 
 | | Crystal structure of murine receptor-interacting protein 4 (Ripk4) D143N bound to TG100-115 | | Descriptor: | 3,3'-(2,4-diaminopteridine-6,7-diyl)diphenol, CHLORIDE ION, Receptor-interacting serine/threonine-protein kinase 4 | | Authors: | Huang, C.S, Hymowitz, S.G. | | Deposit date: | 2017-08-01 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal Structure of Ripk4 Reveals Dimerization-Dependent Kinase Activity.
Structure, 26, 2018
|
|
8XH2
 
 | |
5WNI
 
 | |
6Y1S
 
 | |
7JMY
 
 | |
4PRB
 
 | | Crystal structure of a HLA-B*35:08-HPVG-A4 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRP
 
 | | Crystal structure of TK3 TCR-HLA-B*35:01-HPVG-Q5 complex | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PR5
 
 | | Crystal structure of a HLA-B*35:01-HPVG-D5 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, HLA class I histocompatibility antigen, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRI
 
 | | Crystal structure of TK3 TCR-HLA-B*35:08-HPVG complex | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, Epstein-Barr nuclear antigen 1, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRA
 
 | | Crystal structure of a HLA-B*35:01-HPVG-Q5 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRH
 
 | | Crystal structure of TK3 TCR-HLA-B*35:08-HPVG-D5 complex | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
8IZZ
 
 | |
8BI4
 
 | |
5D2A
 
 | | Bifunctional dendrimers | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Michaud, G, Visini, R, Stocker, A, Darbre, T, Reymond, J.L. | | Deposit date: | 2015-08-05 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Overcoming antibiotic resistance inPseudomonas aeruginosabiofilms using glycopeptide dendrimers.
Chem Sci, 7, 2016
|
|
4S14
 
 | | Crystal structure of the orphan nuclear receptor RORgamma ligand-binding domain in complex with 4alpha-caboxyl, 4beta-methyl-zymosterol (4ACD8) | | Descriptor: | (3beta,4alpha,5beta,14beta)-3-hydroxy-4-methylcholesta-8,24-diene-4-carboxylic acid, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Huang, P, Santori, F.R, Littman, D.R, Rastinejad, F. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-11 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (3.542 Å) | | Cite: | Identification of Natural ROR gamma Ligands that Regulate the Development of Lymphoid Cells.
Cell Metab, 21, 2015
|
|
6BIL
 
 | | HLA-DRB1 in complex with citrullinated fibrinogen peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 74cit69-81, HLA class II histocompatibility antigen, ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-11-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
J. Biol. Chem., 293, 2018
|
|
6BIV
 
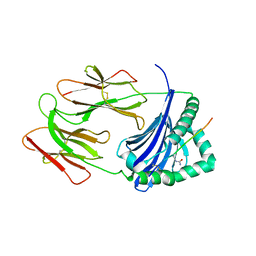 | | HLA-DRB1 in complex with citrullinated LL37 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
J. Biol. Chem., 293, 2018
|
|
6BIX
 
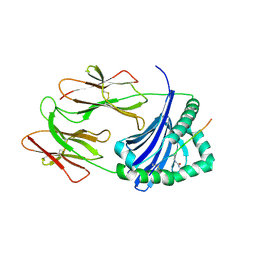 | | HLA-DRB1 in complex with citrullinated LL37 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II DR-beta (HLA-DR B), HLA class II histocompatibility antigen, ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
J. Biol. Chem., 293, 2018
|
|
6BIJ
 
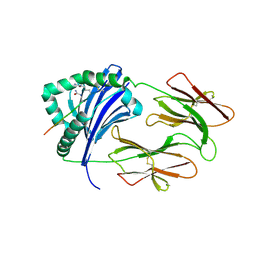 | | HLA-DRB1 in complex with citrullinated fibrinogen peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Citrullinated Fibrinogen 72,74Cit69-81, HLA class II histocompatibility antigen, ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-11-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
J. Biol. Chem., 293, 2018
|
|
6BIY
 
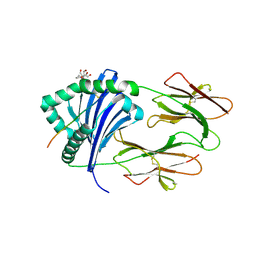 | | HLA-DRB1 in complex with Histone 2B peptide | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II DR-beta (HLA-DR B), ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.050049 Å) | | Cite: | The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
J. Biol. Chem., 293, 2018
|
|
