6FE5
 
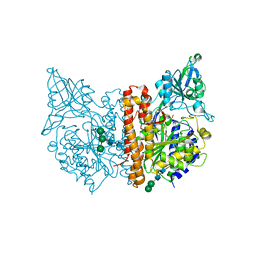 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a inhibitor JHU 2249 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
6M2J
 
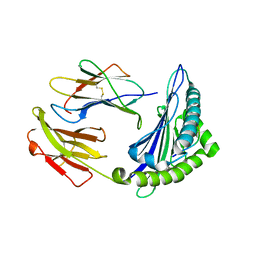 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-1 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
2QFU
 
 | |
3RRC
 
 | |
2Q7S
 
 | |
2GSM
 
 | | Catalytic Core (Subunits I and II) of Cytochrome c oxidase from Rhodobacter sphaeroides | | Descriptor: | CADMIUM ION, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Qin, L, Hiser, C, Mulichak, A, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2C0O
 
 | | Src family kinase Hck with bound inhibitor A-770041 | | Descriptor: | CALCIUM ION, N-(4-{1-[4-(4-ACETYLPIPERAZIN-1-YL)-TRANS-CYCLOHEXYL]-4-AMINO-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL}-2-METHOXYPHENYL)-1-METHYL-1H-INDOLE-2-CARBOXAMIDE, TYROSINE-PROTEIN KINASE HCK | | Authors: | Borhani, D.W, Burchat, A, Calderwood, D.J, Hirst, G.C, Li, B, Loew, A. | | Deposit date: | 2005-09-06 | | Release date: | 2006-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of A-770041, a Src-Family Selective Orally Active Lck Inhibitor that Prevents Organ Allograft Rejection.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4DYG
 
 | | Crystal Structure of a Family GH-19 Chitinase from rye seeds in complex with (GlcNAc)4 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic endochitinase C, ... | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-02-29 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and chitin oligosaccharide-binding mode of a 'loopful' family GH19 chitinase from rye, Secale cereale, seeds
Febs J., 279, 2012
|
|
2PUV
 
 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | 5-AMINO-5-DEOXY-1-O-PHOSPHONO-D-MANNITOL, ACETATE ION, SODIUM ION, ... | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-05-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
3R6Q
 
 | | A triclinic-lattice structure of aspartase from Bacillus sp. YM55-1 | | Descriptor: | Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
2ZRV
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with reduced FMN. | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Isopentenyl-diphosphate delta-isomerase | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
4H1Y
 
 | | Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with PSB11552 | | Descriptor: | 2,2'-[(2-{[2-({[(2S,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)ethyl]amino}-2-oxoethyl)imino]diacetic acid (non-preferred name), 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Pippel, J, Zebisch, M, Knapp, K, Straeter, N. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
1BZM
 
 | |
7FS5
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 17 | | Descriptor: | (10aP)-6-{[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]methyl}-2,3,8,9-tetrahydroxy-5lambda~6~-dibenzo[c,e][1,2]thiazine-5,5(6H)-dione, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FS6
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 18 | | Descriptor: | (10aM)-6-{2-[4-(3,4-dihydroxybenzene-1-sulfonyl)phenyl]ethyl}-2,3,8,9-tetrahydroxy-5lambda~6~-dibenzo[c,e][1,2]thiazine-5,5(6H)-dione, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FRW
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 4 | | Descriptor: | (10aM)-3,8,9-trihydroxy-6H-6lambda~6~-dibenzo[c,e][1,2]oxathiine-6,6-dione, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
7FSD
 
 | | Structure of liver pyruvate kinase in complex with allosteric modulator 44 | | Descriptor: | (1M)-N-({4-[(3,4-dihydroxyphenyl)methyl]phenyl}methyl)-3',4,4',5-tetrahydroxy[1,1'-biphenyl]-2-sulfonamide, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Nilsson, O, Brear, P, Nain-Perez, A, Grotli, M, Hyvonen, M. | | Deposit date: | 2022-12-18 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Tuning liver pyruvate kinase activity up or down with a new class of allosteric modulators.
Eur.J.Med.Chem., 250, 2023
|
|
1OMW
 
 | | Crystal Structure of the complex between G Protein-Coupled Receptor Kinase 2 and Heterotrimeric G Protein beta 1 and gamma 2 subunits | | Descriptor: | G-protein coupled receptor kinase 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-2 subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(T) beta subunit 1 | | Authors: | Lodowski, D.T, Pitcher, J.A, Capel, W.D, Lefkowitz, R.J, Tesmer, J.J.G. | | Deposit date: | 2003-02-26 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Keeping G proteins at Bay: A Complex Between G Protein-Coupled Receptor Kinase 2 and G-Beta-Gamma
Science, 300, 2003
|
|
2GVC
 
 | | Crystal structure of flavin-containing monooxygenase (FMO)from S.pombe and substrate (methimazole) complex | | Descriptor: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1BTW
 
 | |
1UEW
 
 | | Solution Structure of The forth PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein) | | Descriptor: | MEMBRANE ASSOCIATED GUANYLATE KINASE INVERTED-2 (MAGI-2) | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The forth PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein)
To be Published
|
|
2ZRY
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with reduced FMN and IPP. | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, ... | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
7K3L
 
 | |
1P2D
 
 | | Crystal Structure of Glycogen Phosphorylase B in complex with Beta Cyclodextrin | | Descriptor: | Glycogen phosphorylase, muscle form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Pinotsis, N, Leonidas, D.D, Chrysina, E.D, Oikonomakos, N.G, Mavridis, I.M. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The binding of beta- and gamma-cyclodextrins to glycogen phosphorylase b: Kinetic and crystallographic studies.
Protein Sci., 12, 2003
|
|
6FHW
 
 | | Structure of Hormoconis resinae Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
