4PF5
 
 | | Crystal structure of Concanavalin A complexed with a synthetic derivative of high-mannose chain | | Descriptor: | 1,2-ETHANEDIOL, 4-(hydroxymethyl)-1-(alpha-D-mannopyranosyl)-1H-1,2,3-triazole, Concanavalin-A, ... | | Authors: | Lafite, P, Daniellou, R. | | Deposit date: | 2014-04-28 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Synthesis of High-Mannose Oligosaccharide Analogues through Click Chemistry: True Functional Mimics of Their Natural Counterparts Against Lectins?
Chemistry, 21, 2015
|
|
3QQ7
 
 | |
6S2H
 
 | |
3QQ0
 
 | | Crystal structure of a deletion mutant (N59) of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Jameson, G.B, Parker, E.J. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting the role of a key conserved motif for substrate selection and catalysis by 3-deoxy-D-manno-octulosonate 8-phosphate synthase
Biochemistry, 50, 2011
|
|
5YQ7
 
 | | Cryo-EM structure of the RC-LH core complex from Roseiflexus castenholzii | | Descriptor: | 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, BACTERIOCHLOROPHYLL A, ... | | Authors: | Shi, Y, Xin, Y.Y, Niu, T.X, Wang, Q.Q, Niu, W.Q, Huang, X.J, Ding, W, Blankenship, R.E, Xu, X.L, Sun, F. | | Deposit date: | 2017-11-05 | | Release date: | 2018-05-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote.
Nat Commun, 9, 2018
|
|
4OOA
 
 | | CRYSTAL STRUCTURE of NAF1 (MINER1): H114C THE REDOX-ACTIVE 2FE-2S PROTEIN | | Descriptor: | CDGSH iron-sulfur domain-containing protein 2, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Tamir, S, Eisenberg-Domovich, Y, Conlan, A.R, Stofleth, J.T, Lipper, C.H, Paddock, M.L, Mittler, R, Jennings, P.A, Livnah, O, Nechushtai, R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A point mutation in the [2Fe-2S] cluster binding region of the NAF-1 protein (H114C) dramatically hinders the cluster donor properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4F9M
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | (9aS)-8-acetyl-N-[(2-ethyl-4-fluoronaphthalen-1-yl)methyl]-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peroxisome proliferator-activated receptor gamma, peptide from Nuclear receptor coactivator 1 | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2012-05-19 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and biological evaluation of novel (-)-cercosporamide derivatives as potent selective PPARg modulators
Eur.J.Med.Chem., 54, 2012
|
|
1ZI9
 
 | | Crystal Structure Analysis of the dienelactone hydrolase (E36D, C123S) mutant- 1.5 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZJ4
 
 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6LB4
 
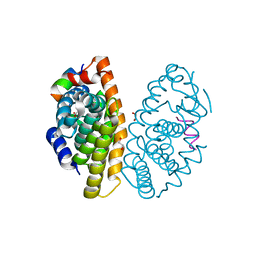 | | Crystal structure of dimeric RXR-LBD complexed with NEt-3ME and TIF2 co-activator | | Descriptor: | 1,2-ETHANEDIOL, 6-[ethyl-[3-(2-methoxyethoxy)-4-propan-2-yl-phenyl]amino]pyridine-3-carboxylic acid, Nuclear receptor coactivator 2, ... | | Authors: | Imai, D, Numoto, N, Nakano, S, Kakuta, H, Ito, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dimeric RXR-LBD complexed with NEt-3ME and TIF2 co-activator
To Be Published
|
|
7FCY
 
 | | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor | | Descriptor: | (1S)-2-methyl-1-(2-methyl-1,2,4-triazol-3-yl)propan-1-amine, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor
To Be Published
|
|
1ZIX
 
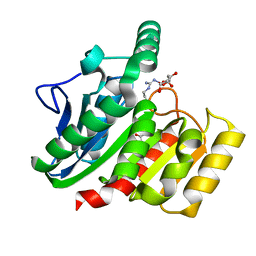 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, R105H, C123S, G211D, K234N)- 1.8 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3AI5
 
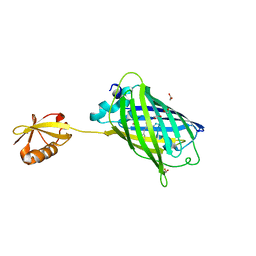 | | Crystal structure of yeast enhanced green fluorescent protein-ubiquitin fusion protein | | Descriptor: | 1,2-ETHANEDIOL, yeast enhanced green fluorescent protein,Ubiquitin | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1UYP
 
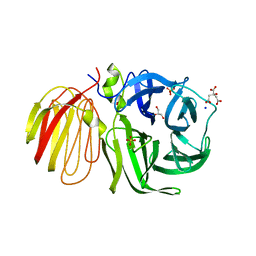 | | The three-dimensional structure of beta-fructosidase (invertase) from Thermotoga maritima | | Descriptor: | BETA-FRUCTOSIDASE, CITRIC ACID, GLYCEROL, ... | | Authors: | Alberto, F, Bignon, C, Sulzenbacher, G, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-02 | | Release date: | 2004-03-22 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of invertase (beta-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases.
J. Biol. Chem., 279, 2004
|
|
4RN0
 
 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3,17-dithia-7,10,14,19,20-pentaazatricyclo[14.2.1.1~2,5~]icosa-1(18),2(20),16(19)-triene-6,9,13-trione, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
2Q9K
 
 | |
4IZZ
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 21mer DNA target | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*C)-3'), SULFATE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3MSD
 
 | | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. | | Descriptor: | ACETATE ION, GLYCEROL, Intra-cellular xylanase ixt6, ... | | Authors: | Solomon, V, Zolotnitsky, G, Alhadeff, R, Shoham, Y, Shoham, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus.
TO BE PUBLISHED
|
|
1V2K
 
 | | Factor XA specific Inhibitor in complex with bovine trypsin variant X(triple.Glu)bT.D2 | | Descriptor: | CALCIUM ION, SULFATE ION, Trypsin, ... | | Authors: | Rauh, D, Klebe, G, Stubbs, M.T. | | Deposit date: | 2003-10-17 | | Release date: | 2004-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding protein-ligand interactions: the price of protein flexibility
J.Mol.Biol., 335, 2004
|
|
3MKS
 
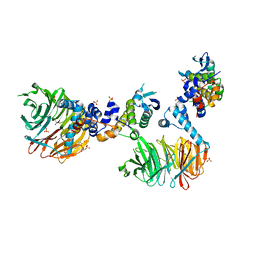 | | Crystal Structure of yeast Cdc4/Skp1 in complex with an allosteric inhibitor SCF-I2 | | Descriptor: | 1,1'-binaphthalene-2,2'-dicarboxylic acid, Cell division control protein 4, GLYCEROL, ... | | Authors: | Orlicky, S, Sicheri, F, Tyers, M, Tang, X. | | Deposit date: | 2010-04-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase.
Nat.Biotechnol., 28, 2010
|
|
2Q61
 
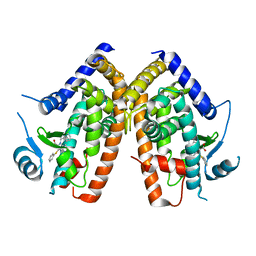 | |
5TQG
 
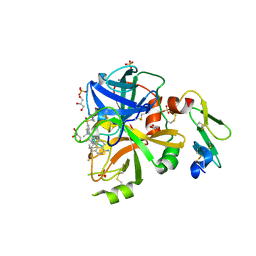 | | Factor VIIa in complex with the inhibitor (5R,11R)-11-[(1-amino-4-fluoroisoquinolin-6-yl)amino]-16-(cyclopropylsulfonyl)-7-(2,2-difluoroethoxy)-5,13-dimethyl-2,13-diazatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaene-3,12-dione | | Descriptor: | (5R,11R)-11-[(1-amino-4-fluoroisoquinolin-6-yl)amino]-16-(cyclopropylsulfonyl)-7-(2,2-difluoroethoxy)-5,13-dimethyl-2,13-diazatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaene-3,12-dione, CALCIUM ION, Factor VIIa (Heavy Chain), ... | | Authors: | Wei, A. | | Deposit date: | 2016-10-24 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Novel Meta-Linked Phenylglycine Macrocyclic FVIIa Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
7EVO
 
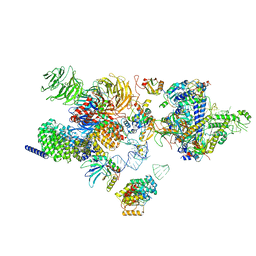 | | The cryo-EM structure of the human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, RNA helicase, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
1Z2U
 
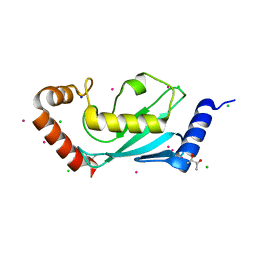 | | The 1.1A crystallographic structure of ubiquitin-conjugating enzyme (ubc-2) from Caenorhabditis elegans: functional and evolutionary significance | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gavira, J.A, DiGiamamarino, E, Tempel, W, Liu, Z.J, Wang, B.C, Meehan, E, Ng, J.D, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1A crystallographic structure of ubiquitin-conjugating enzyme (ubc-2) from Caenorhabditis elegans: functional and evolutionary significance
To be published
|
|
4TOH
 
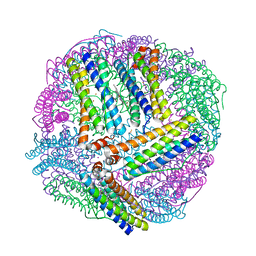 | | 1.80A resolution structure of Iron Bound BfrB (C89S, K96C) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
