2N0I
 
 | | NMR solution structure for di-sulfide 11mer peptide | | Descriptor: | di-sulfide 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N09
 
 | | NMR structure of a short hydrophobic 11mer peptide in DMSO-d6/H2O (1:3) solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2015-05-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N7O
 
 | | NMR Structure of Peptide PG-990 in DPC micelles | | Descriptor: | Peptide PG-990 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
2N7T
 
 | | NMR structure of Peptide PG-992 in DPC micelles | | Descriptor: | Peptide PG-992 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
2N7N
 
 | | NMR structure of Peptide PG-989 in DPC micelles | | Descriptor: | Peptide PG-989 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
3DJP
 
 | | Bovine Seminal Ribonuclease- Uridine 3' phosphate complex | | Descriptor: | Seminal ribonuclease, URACIL ARABINOSE-3'-PHOSPHATE | | Authors: | Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the ribonucleolytic active site of bovine seminal ribonuclease. The binding of pyrimidinyl phosphonucleotide inhibitors
Eur.J.Med.Chem., 44, 2009
|
|
3R4N
 
 | | Optimization of Potent, Selective, and Orally Bioavailable Pyrrolodinopyrimidine-containing Inhibitors of Heat Shock Protein 90. Identification of Development Candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide | | Descriptor: | 4-[2-chloro-6-(4,4,4-trifluorobutoxy)phenyl]-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Gajiwala, K.S. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. Identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide.
J.Med.Chem., 54, 2011
|
|
3R4O
 
 | | Optimization of Potent, Selective, and Orally Bioavailable Pyrrolodinopyrimidine-containing Inhibitors of Heat Shock Protein 90. Identification of Development Candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide | | Descriptor: | 2-amino-N-cyclobutyl-4-[2,4-dichloro-6-(4,4,4-trifluorobutoxy)phenyl]-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Gajiwala, K.S. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. Identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide.
J.Med.Chem., 54, 2011
|
|
3R4P
 
 | | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide | | Descriptor: | 2-amino-4-{2,4-dichloro-6-[2-(1H-pyrazol-1-yl)ethoxy]phenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Gajiwala, K.S. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. Identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide.
J.Med.Chem., 54, 2011
|
|
2ZL9
 
 | | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation and crystal structure | | Descriptor: | (1R,2R,3R,5Z)-17-{(1S)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethoxy)-9,10-secoestra-5,7,16-triene-1,3-diol, Coactivator peptide DRIP, Vitamin D3 receptor | | Authors: | Shimizu, M, Miyamoto, Y, Nakabayashi, M, Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure
Bioorg.Med.Chem., 16, 2008
|
|
2ZLC
 
 | | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation and crystal structure | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Coactivator peptide DRIP, Vitamin D3 receptor | | Authors: | Shimizu, M, Miyamoto, Y, Nakabayashi, M, Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure
Bioorg.Med.Chem., 16, 2008
|
|
3AKO
 
 | | Crystal Structure of the Reassembled Venus | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, Venus | | Authors: | Isogai, M, Tada, T. | | Deposit date: | 2010-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and characteristics of reassembled fluorescent protein, a new insight into the reassembly mechanisms
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3DJQ
 
 | | Bovine Seminal Ribonuclease- Uridine 5' diphosphate complex | | Descriptor: | Seminal ribonuclease, URIDINE-5'-DIPHOSPHATE | | Authors: | Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Mapping the ribonucleolytic active site of bovine seminal ribonuclease. The binding of pyrimidinyl phosphonucleotide inhibitors
Eur.J.Med.Chem., 44, 2009
|
|
3DJV
 
 | | Bovine Seminal Ribonuclease- cytidine 3' phosphate complex | | Descriptor: | CYTIDINE-3'-MONOPHOSPHATE, Seminal ribonuclease | | Authors: | Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the ribonucleolytic active site of bovine seminal ribonuclease. The binding of pyrimidinyl phosphonucleotide inhibitors
Eur.J.Med.Chem., 44, 2009
|
|
7KZI
 
 | | Intermediate state (QQQ) of near full-length DnaK alternatively fused with a substrate peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chaperone protein DnaK fused with substrate peptide,Chaperone protein DnaK fused with substrate peptide, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Intermediates in allosteric equilibria of DnaK-ATP interactions with substrate peptides
Acta Crystallogr.,Sect.D, 77, 2021
|
|
3I25
 
 | | Potent Beta-Secretase 1 hydroxyethylene Inhibitor | | Descriptor: | Beta-secretase 1, N-[(2S,3S,5R)-1-(3,5-difluorophenoxy)-3-hydroxy-5-(2-methoxyethoxy)-6-[[(2S)-3-methyl-1-oxo-1-(phenylmethylamino)butan-2-yl]amino]-6-oxo-hexan-2-yl]-5-(methyl-methylsulfonyl-amino)-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Lindberg, J.D, Borkakoti, N, Nystrom, S. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of potent BACE-1 inhibitors containing a new hydroxyethylene (HE) scaffold: exploration of P1' alkoxy residues and an aminoethylene (AE) central core
Bioorg.Med.Chem., 18, 2010
|
|
3TYV
 
 | |
3RAS
 
 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with a lipophilic phosphonate inhibitor | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-(N-HYDROXYACETAMIDO)-1-(3,4-DICHLOROPHENYL)PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Diao, J, Deng, L, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase by lipophilic phosphonates: SAR, QSAR, and crystallographic studies.
J.Med.Chem., 54, 2011
|
|
2ZLA
 
 | | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation and crystal structure | | Descriptor: | (1R,2S,3R,5Z,7E)-17-{(1R)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethoxy)-9,10-secoestra-5,7,16-triene-1,3-diol, Coactivator peptide DRIP, Vitamin D3 receptor | | Authors: | Shimizu, M, Miyamoto, Y, Nakabayashi, M, Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure
Bioorg.Med.Chem., 16, 2008
|
|
3DJX
 
 | | Bovine Seminal Ribonuclease- cytidine 5' phosphate complex | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Seminal ribonuclease | | Authors: | Dossi, K, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-06-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Mapping the ribonucleolytic active site of bovine seminal ribonuclease. The binding of pyrimidinyl phosphonucleotide inhibitors
Eur.J.Med.Chem., 44, 2009
|
|
3UIC
 
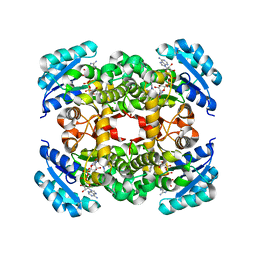 | | Crystal Structure of FabI, an Enoyl Reductase from F. tularensis, in complex with a Novel and Potent Inhibitor | | Descriptor: | 1-(3,4-dichlorobenzyl)-5,6-dimethyl-1H-benzimidazole, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mehboob, S, Santarsiero, B.D, Truong, K, Johnson, M.E. | | Deposit date: | 2011-11-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and enzymatic analyses reveal the binding mode of a novel series of Francisella tularensis enoyl reductase (FabI) inhibitors.
J.Med.Chem., 55, 2012
|
|
1YL8
 
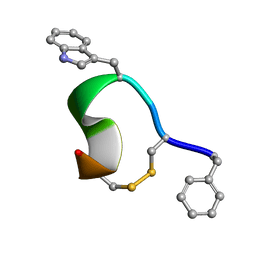 | | 3D Solution Structure of [Tyr3]Octreotate derivatives in DMSO | | Descriptor: | [Tyr3]Octreotate peptide | | Authors: | Spyroulias, G.A, Galanis, A.S, Petrou, C, Vahliotis, D, Sotiriou, P, Nikolopoulou, A, Nock, B, Maina, T, Cordopatis, P. | | Deposit date: | 2005-01-19 | | Release date: | 2005-09-20 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 3D solution structure of [Tyr3]octreotate derivatives in DMSO: structure differentiation of peptide core due to chelate group attachment and biologically active conformation.
Med.Chem., 1, 2005
|
|
1YL9
 
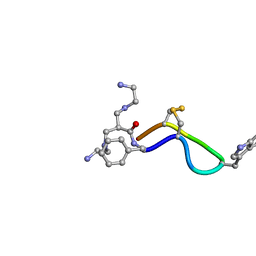 | | 3D Solution Structure of [Tyr3]Octreotate derivatives in DMSO | | Descriptor: | 3-[(2-AMINOETHYL)AMINO]-2-{[(2-AMINOETHYL)AMINO]METHYL}PROPANAL, [Tyr3]Octreotate | | Authors: | Spyroulias, G.A, Galanis, A.S, Petrou, C, Vahliotis, D, Sotiriou, P, Nikolopoulou, A, Nock, B, Maina, T, Cordopatis, P. | | Deposit date: | 2005-01-19 | | Release date: | 2005-09-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | 3D solution structure of [Tyr3]octreotate derivatives in DMSO: structure differentiation of peptide core due to chelate group attachment and biologically active conformation.
Med.Chem., 1, 2005
|
|
3M2N
 
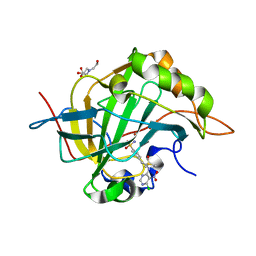 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{2-[N-(6-chloro-5-nitropyrimidin-4-yl)amino]ethyl}benzenesulfonamide | | Descriptor: | 4-{2-[(6-chloro-5-nitropyrimidin-4-yl)amino]ethyl}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-08 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
6HVH
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 1 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-(oxan-4-yl)pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Jakubiec, K, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and Structure-Activity Relationships of N-Aryl 6-Aminoquinoxalines as Potent PFKFB3 Kinase Inhibitors.
ChemMedChem, 14, 2019
|
|
