1XEY
 
 | | Crystal structure of the complex of Escherichia coli GADA with glutarate at 2.05 A resolution | | Descriptor: | ACETATE ION, GLUTARIC ACID, Glutamate decarboxylase alpha, ... | | Authors: | Dutyshev, D.I, Darii, E.L, Fomenkova, N.P, Pechik, I.V, Polyakov, K.M, Nikonov, S.V, Andreeva, N.S, Sukhareva, B.S. | | Deposit date: | 2004-09-13 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Escherichia coli glutamate decarboxylase (GADalpha) in complex with glutarate at 2.05 angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3CII
 
 | | Structure of NKG2A/CD94 bound to HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen peptide, HLA class I histocompatibility antigen, ... | | Authors: | Strong, R.K, Kaiser, B.K, Pizarro, J.C. | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.41 Å) | | Cite: | Structural basis for NKG2A/CD94 recognition of HLA-E.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1CMU
 
 | | THE ROLE OF ASPARTATE-235 IN THE BINDING OF CATIONS TO AN ARTIFICIAL CAVITY AT THE RADICAL SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Trester, M.L, Jensen, G.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1995-04-10 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of aspartate-235 in the binding of cations to an artificial cavity at the radical site of cytochrome c peroxidase.
Protein Sci., 4, 1995
|
|
2DKN
 
 | |
1R41
 
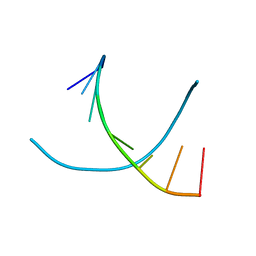 | | Crystal structures of d(Gm5CGm5CGCGC) and d(GCGCGm5CGm5C): Effects of methylation on alternating DNA octamers | | Descriptor: | 5'-D(*GP*CP*GP*CP*GP*(5CM)P*GP*(5CM))-3' | | Authors: | Shi, K, Pan, B, Tippin, D, Sundaralingam, M. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of d(Gm5)CGm5CGCGC) and d(GCGCGm5CGm5C): effects of methylation on alternating DNA octamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SGV
 
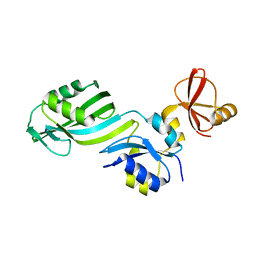 | | STRUCTURE OF TRNA PSI55 PSEUDOURIDINE SYNTHASE (TRUB) | | Descriptor: | tRNA pseudouridine synthase B | | Authors: | Chaudhuri, B.N, Chan, S, Perry, L.J, Yeates, T.O, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the apo forms of psi 55 tRNA pseudouridine synthase from Mycobacterium tuberculosis: a hinge at the base of the catalytic cleft.
J.Biol.Chem., 279, 2004
|
|
1CKO
 
 | |
1SM2
 
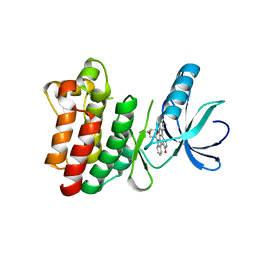 | | Crystal structure of the phosphorylated Interleukin-2 tyrosine kinase catalytic domain | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C.M, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-08 | | Release date: | 2004-07-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
2DBV
 
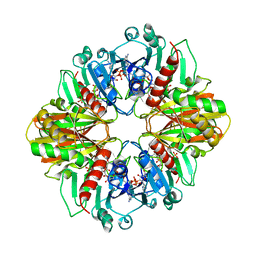 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH ASP 32 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1996-12-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
1RXP
 
 | |
3HJT
 
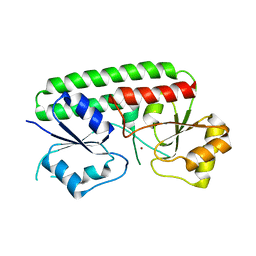 | |
3URE
 
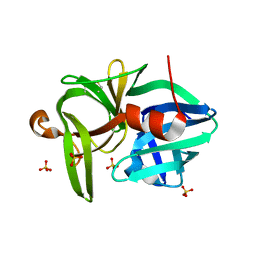 | |
1AGS
 
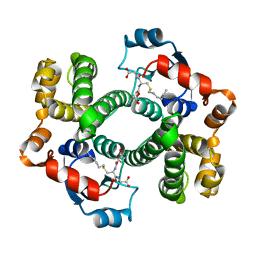 | |
1ESB
 
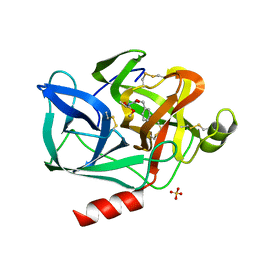 | | DIRECT STRUCTURE OBSERVATION OF AN ACYL-ENZYME INTERMEDIATE IN THE HYDROLYSIS OF AN ESTER SUBSTRATE BY ELASTASE | | Descriptor: | CALCIUM ION, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Ding, X, Rasmussen, B, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-04 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct structural observation of an acyl-enzyme intermediate in the hydrolysis of an ester substrate by elastase.
Biochemistry, 33, 1994
|
|
2LEG
 
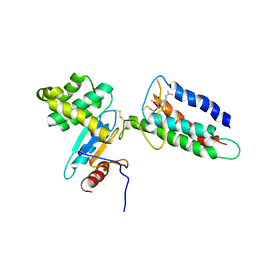 | | Membrane protein complex DsbB-DsbA structure by joint calculations with solid-state NMR and X-ray experimental data | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein DsbA, UBIQUINONE-1, ... | | Authors: | Tang, M, Sperling, L.J, Berthold, D.A, Schwieters, C.D, Nesbitt, A.E, Nieuwkoop, A.J, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-26 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data.
J.Biomol.Nmr, 51, 2011
|
|
2L1Z
 
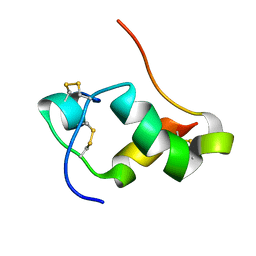 | | NMR Structure of human insulin mutant GLY-B20-D-ALA, GLY-B23-D-ALA PRO-B28-LYS, LYS-B29-PRO, 20 Structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wan, Z.L, Hua, Q.X, Huang, K, Hu, S.Q, Philips, N.B, Katsoyannis, J.W, Weiss, M.A. | | Deposit date: | 2010-08-09 | | Release date: | 2011-08-31 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Chiral Protein Engineering and its Application in G Health
To be Published
|
|
2HCI
 
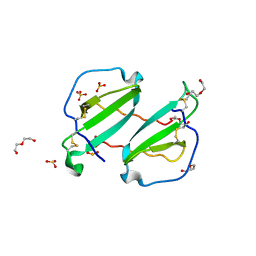 | | Structure of Human Mip-3a Chemokine | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, SULFITE ION, ... | | Authors: | Malik, Z.A, Tack, B.F. | | Deposit date: | 2006-06-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of human MIP-3alpha chemokine.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2KE5
 
 | | Solution structure and dynamics of the small GTPase Ralb in its active conformation: significance for effector protein binding | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Ral-B | | Authors: | Fenwick, R, Prasannan, S, Campbell, L.J, Nietlispach, D, Evetts, K.A, Camonis, J, Mott, H.R, Owen, D. | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the small GTPase RalB in its active conformation: significance for effector protein binding
Biochemistry, 48, 2009
|
|
2M1T
 
 | |
3QIN
 
 | |
1IPK
 
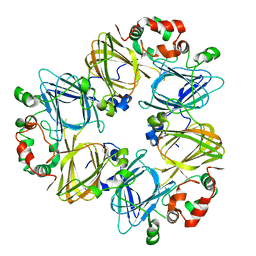 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS | | Descriptor: | BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
4DB4
 
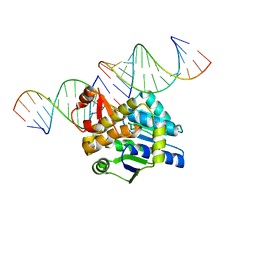 | | Mss116p DEAD-box helicase domain 2 bound to a chimaeric RNA-DNA duplex | | Descriptor: | 5'-R(*GP*GP*GP*CP*GP*GP*G)-D(P*CP*CP*CP*GP*CP*CP*C)-3', ATP-dependent RNA helicase MSS116, mitochondrial | | Authors: | Mallam, A.L, Del Campo, M, Gilman, B.D, Sidote, D.J, Lambowitz, A. | | Deposit date: | 2012-01-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.599 Å) | | Cite: | Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p.
Nature, 490, 2012
|
|
4DCU
 
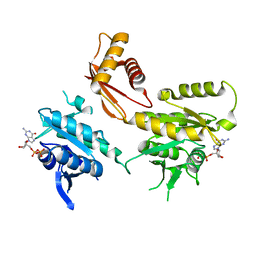 | | Crystal Structure of B. subtilis EngA in complex with GDP | | Descriptor: | GTP-BINDING PROTEIN ENGA, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Reiser, J.-B, Housset, D, Foucher, A.-E, Jault, J.-M. | | Deposit date: | 2012-01-18 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potassium Acts as a GTPase-Activating Element on Each Nucleotide-Binding Domain of the Essential Bacillus subtilis EngA.
Plos One, 7, 2012
|
|
1TLV
 
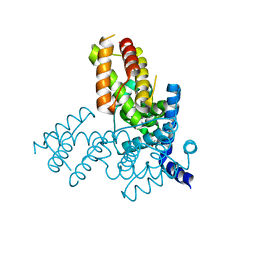 | | Structure of the native and inactive LicT PRD from B. subtilis | | Descriptor: | Transcription antiterminator licT | | Authors: | Graille, M, Zhou, C.-Z, Receveur-Brechot, V, Collinet, B, Declerck, N, van Tilbeurgh, H. | | Deposit date: | 2004-06-10 | | Release date: | 2005-02-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Activation of the LicT Transcriptional Antiterminator Involves a Domain Swing/Lock Mechanism Provoking Massive Structural Changes
J.Biol.Chem., 280, 2005
|
|
3VAP
 
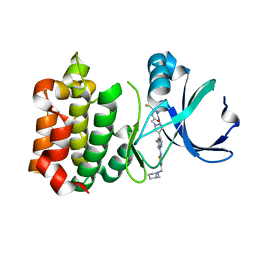 | | Synthesis and SAR Studies of imidazo-[1,2-a]-pyrazine Aurora kinase inhibitors with improved off target kinase selectivity | | Descriptor: | 3-(1-{2-[(3-fluoropyridinium-4-yl)amino]-2-oxoethyl}-1H-pyrazol-4-yl)-6-methyl-8-[(3-{[(1R,3R)-3-methylpiperidinium-1-yl]methyl}-1,2-thiazol-5-yl)amino]imidazo[1,2-a]pyrazin-1-ium, Aurora kinase A | | Authors: | Voss, M.E, Rainka, M.P, Fleming, M, Peterson, L.H, Belanger, D.B, Siddiqui, M.A, Hruza, A, Voigt, J, Basso, A.D, Gray, K. | | Deposit date: | 2011-12-29 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Synthesis and SAR studies of imidazo-[1,2-a]-pyrazine Aurora kinase inhibitors with improved off-target kinase selectivity.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
