3WJX
 
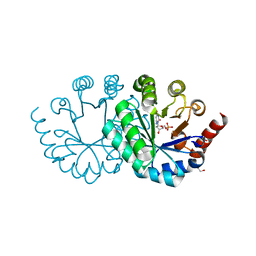 | | Wild-type orotidine 5'-monophosphate decarboxylase from M. thermoautotrophicus complexed with 6-amino-UMP | | Descriptor: | 6-AMINOURIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fujihashi, M, Kuroda, S, Pai, E.F, Miki, K. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Substrate distortion contributes to the catalysis of orotidine 5'-monophosphate decarboxylase.
J.Am.Chem.Soc., 135, 2013
|
|
1I36
 
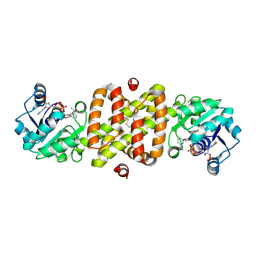 | | Structure of Conserved Protein MTH1747 of Unknown Function Reveals Structural Similarity with 3-Hydroxyacid Dehydrogenases | | Descriptor: | CONSERVED HYPOTHETICAL PROTEIN MTH1747, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korolev, S.V, Dementieva, I.S, Christendat, D, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-02-13 | | Release date: | 2002-05-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURAL SIMILARITIES OF MTH1747 HYPOTHETICAL PROTEIN FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH 3-HYDROXYACID DEHYDROGENASES
to be published
|
|
2MMZ
 
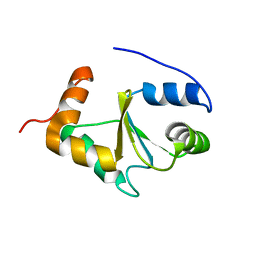 | | Solution structure of the apo form of human glutaredoxin 5 | | Descriptor: | Glutaredoxin-related protein 5, mitochondrial | | Authors: | Banci, L, Brancaccio, D, Ciofi-Baffoni, S, Del Conte, R, Gadepalli, R, Mikolajczyk, M, Neri, S, Piccioli, M, Winkelmann, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3IL1
 
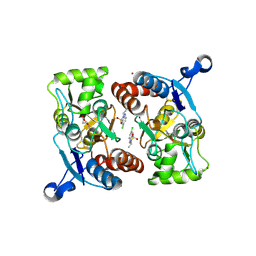 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, IDRA-21 | | Descriptor: | (3S)-7-chloro-3-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-06 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3ILT
 
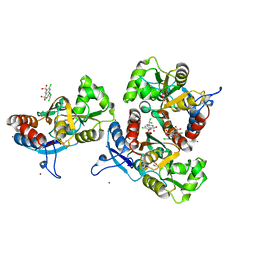 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, trichlormethiazide | | Descriptor: | 6-CHLORO-3-(DICHLOROMETHYL)-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE-7-SULFONAMIDE 1,1-DIOXIDE, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IK6
 
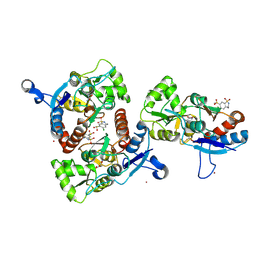 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, chlorothiazide | | Descriptor: | 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
2KB9
 
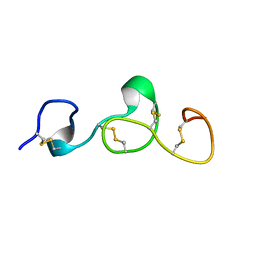 | |
3IJO
 
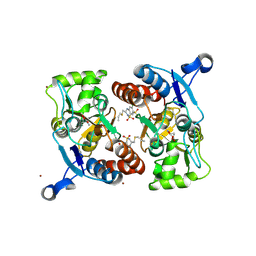 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, althiazide | | Descriptor: | (3S)-6-chloro-3-[(prop-2-en-1-ylsulfanyl)methyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IJX
 
 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydrochlorothiazide | | Descriptor: | 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3ILU
 
 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydroflumethiazide | | Descriptor: | 6-(trifluoromethyl)-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3LDC
 
 | | High resolution open MthK pore structure crystallized in 100 mM K+ | | Descriptor: | Calcium-gated potassium channel mthK, POTASSIUM ION | | Authors: | Ye, S. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel insights into K(+) selectivity from high-resolution structures of an open K(+) channel pore.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1MRO
 
 | | METHYL-COENZYME M REDUCTASE | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | Authors: | Ermler, U, Grabarse, W. | | Deposit date: | 1997-10-01 | | Release date: | 1998-11-11 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation.
Science, 278, 1997
|
|
2GAG
 
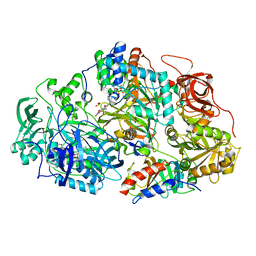 | | Heteroteterameric sarcosine: structure of a diflavin metaloenzyme at 1.85 a resolution | | Descriptor: | 2-FUROIC ACID, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, Z.W, Hassan-Abdulah, A, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Heterotetrameric sarcosine oxidase: structure of a diflavin metalloenzyme at 1.85 a resolution.
J.Mol.Biol., 360, 2006
|
|
2GAH
 
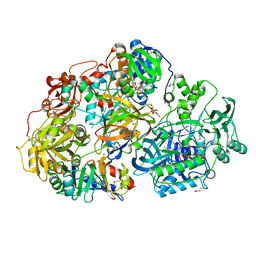 | | Heterotetrameric sarcosine: structure of a diflavin metaloenzyme at 1.85 a resolution | | Descriptor: | 2-FUROIC ACID, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, Z.W, Hassan-Abdulah, A, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Heterotetrameric sarcosine oxidase: structure of a diflavin metalloenzyme at 1.85 a resolution.
J.Mol.Biol., 360, 2006
|
|
1KJN
 
 | |
1LOJ
 
 | | Crystal structure of a Methanobacterial Sm-like archaeal protein (SmAP1) bound to uridine-5'-monophosphate (UMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, URIDINE, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Mura, C, Kozhukhovsky, A, Eisenberg, D. | | Deposit date: | 2002-05-06 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs)
Protein Sci., 12, 2003
|
|
1ZPS
 
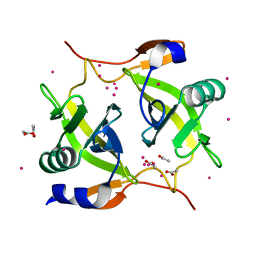 | | Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI | | Descriptor: | ACETIC ACID, CADMIUM ION, Phosphoribosyl-AMP cyclohydrolase | | Authors: | Sivaraman, J, Myers, R.S, Boju, L, Sulea, T, Cygler, M, Davisson, V.J, Schrag, J.D, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Methanobacterium thermoautotrophicum Phosphoribosyl-AMP Cyclohydrolase HisI.
Biochemistry, 44, 2005
|
|
7C8M
 
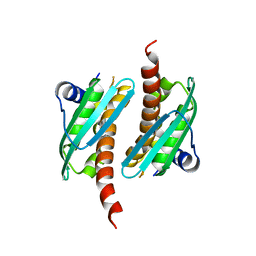 | | Crystal structure of IscU wild-type | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Nitrogen-fixing NifU domain protein | | Authors: | Kunichika, K, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure of the Dimeric State of IscU Harboring Two Adjacent [2Fe-2S] Clusters Provides Mechanistic Insights into Cluster Conversion to [4Fe-4S].
Biochemistry, 60, 2021
|
|
7C8N
 
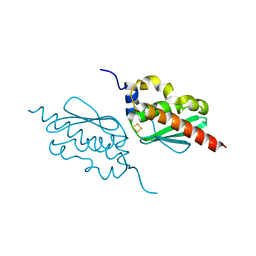 | | Crystal structure of IscU H106A variant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Nitrogen-fixing NifU domain protein | | Authors: | Kunichika, K, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Dimeric State of IscU Harboring Two Adjacent [2Fe-2S] Clusters Provides Mechanistic Insights into Cluster Conversion to [4Fe-4S].
Biochemistry, 60, 2021
|
|
7C8O
 
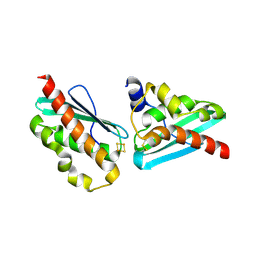 | |
7CNV
 
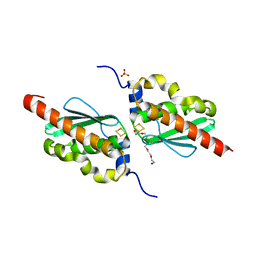 | |
1JRI
 
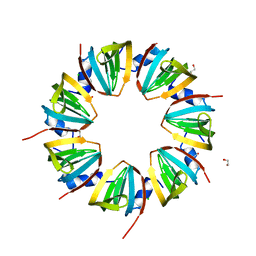 | |
2PTF
 
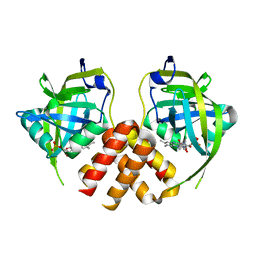 | | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Uncharacterized protein MTH_863 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN.
To be Published
|
|
2PMR
 
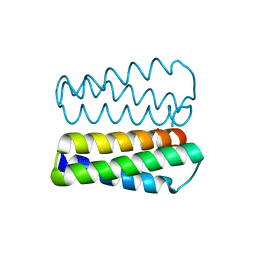 | | Crystal structure of a protein of unknown function from Methanobacterium thermoautotrophicum | | Descriptor: | Uncharacterized protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structure of a protein of unknown function from Methanobacterium thermoautotrophicum.
To be Published
|
|
2OGU
 
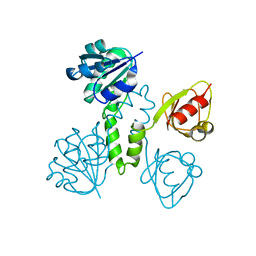 | |
