4JP4
 
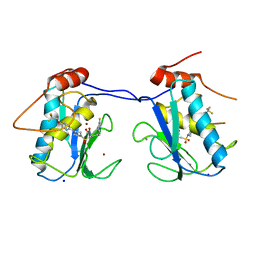 | | Mmp13 in complex with a reverse hydroxamate Zn-binder | | Descriptor: | CALCIUM ION, Collagenase 3, N-[(2S)-4-(5-fluoropyrimidin-2-yl)-1-({4-[5-(2,2,2-trifluoroethoxy)pyrimidin-2-yl]piperazin-1-yl}sulfonyl)butan-2-yl]-N-hydroxyformamide, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydantoin based inhibitors of MMP13--discovery of AZD6605.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2ANQ
 
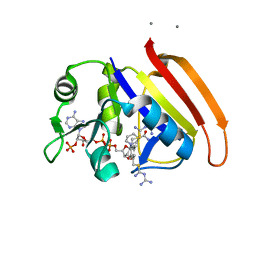 | | Crystal Structure of E.coli DHFR in complex with NADPH and the inhibitor compound 10a. | | Descriptor: | (2,5-dimethylbenzene-1,4-diyl)dimethanediyl bis(N-carbamimidoylcarbamimidothioate), Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
2ANO
 
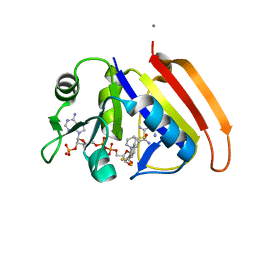 | | Crystal structure of E.coli dihydrofolate reductase in complex with NADPH and the inhibitor MS-SH08-17 | | Descriptor: | 1-{[N-(1-IMINO-GUANIDINO-METHYL)]SULFANYLMETHYL}-3-TRIFLUOROMETHYL-BENZENE, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
4DRQ
 
 | | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-1-((S)-1-(3,5-dichlorophenylsulfonyl)piperidine-2-carbonyloxy)-3-(3,4-dimethoxy -phenyl)propyl)phenoxy)acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1S)-1-[({(2S)-1-[(3,5-dichlorophenyl)sulfonyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Wang, Y, Hoogeland, B, Bracher, A, Hausch, F, Schneider, S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
3I1Y
 
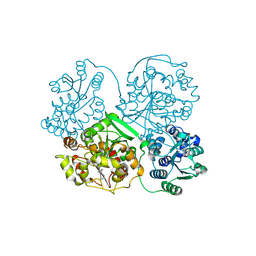 | | Crystal Structure of soluble epoxide Hydrolase | | Descriptor: | Epoxide hydrolase 2, N-(3,3-diphenylpropyl)pyridine-3-carboxamide | | Authors: | Farrow, N.A. | | Deposit date: | 2009-06-28 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Structure-based optimization of arylamides as inhibitors of soluble epoxide hydrolase.
J.Med.Chem., 52, 2009
|
|
1T8D
 
 | | Structure of the C-type lectin domain of CD23 | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor | | Authors: | Hibbert, R.G, Teriete, P, Grundy, G.J, Sutton, B.J, Gould, H.J, McDonnell, J.M. | | Deposit date: | 2004-05-12 | | Release date: | 2005-07-26 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The structure of human CD23 and its interactions with IgE and CD21
J.Exp.Med., 202, 2005
|
|
2J2U
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5-CHLORO-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-5-CHLORO-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-SULFONAMIDE, COAGULATION FACTOR X HEAVY CHAIN, COAGULATION FACTOR X LIGHT CHAIN | | Authors: | Senger, S, Convery, M.A, Chan, C, Watson, N.S. | | Deposit date: | 2006-08-17 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arylsulfonamides: A Study of the Relationship between Activity and Conformational Preferences for a Series of Factor Xa Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1XEP
 
 | | Catechol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | BETA-MERCAPTOETHANOL, CATECHOL, Lysozyme, ... | | Authors: | Graves, A.P, Brenk, R, Shoichet, B.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Decoys for docking.
J.Med.Chem., 48, 2005
|
|
3IEO
 
 | | The coumarin-binding site in carbonic anhydrase: the antiepileptic lacosamide as an example | | Descriptor: | BENZOIC ACID, Carbonic anhydrase 2, MERCURY (II) ION, ... | | Authors: | Temperini, C, Innocenti, A, Scozzafava, A, Parkkila, S, Supuran, C.T. | | Deposit date: | 2009-07-23 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The coumarin-binding site in carbonic anhydrase accommodates structurally diverse inhibitors: the antiepileptic lacosamide as an example and lead molecule for novel classes of carbonic anhydrase inhibitors
J.Med.Chem., 53, 2010
|
|
6KE0
 
 | | Crystal structure of PDE10A in complex with a triazolopyrimidine inhibitor | | Descriptor: | 2-(5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)-1-[(2S)-2-methyl-1,2-dihydroimidazo[1,2-a]benzimidazol-3-yl]ethanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis, SAR study, and biological evaluation of novel 2,3-dihydro-1H-imidazo[1,2-a]benzimidazole derivatives as phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
6KDX
 
 | | Crystal structure of PDE10A in complex with a triazolopyrimidine inhibitor | | Descriptor: | MAGNESIUM ION, N-[2-(5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]quinolin-2-amine, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Synthesis, SAR study, and biological evaluation of novel 2,3-dihydro-1H-imidazo[1,2-a]benzimidazole derivatives as phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
1ZRB
 
 | | Thrombin in complex with an azafluorenyl inhibitor 23b | | Descriptor: | 11-peptide hirudin fragment, 3-AZA-9-HYDROXY-9-FLUORENYLCARBONYL-L-PROLYL-2-AMINOMETHYL-5-CHLOROBENZYLAMIDE, N-OXIDE, ... | | Authors: | Stauffer, K.J, Williams, P.D, Selnick, H.G, Nantermet, P.G, Newton, C.L, Homnick, C.F, Zrada, M.M, Lewis, S.D, Lucas, B.J, Krueger, J.A, Pietrak, B.L, Lyle, E.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Wallace, A.A, Sitko, G.R, Cook, J.J, Holahan, M.A, Stranieri-Michener, M, Leonard, Y.M, Lynch Jr, J.J, McMasters, D.R, Yan, Y. | | Deposit date: | 2005-05-19 | | Release date: | 2005-06-07 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 9-hydroxyazafluorenes and their use in thrombin inhibitors
J.Med.Chem., 48, 2005
|
|
2HWH
 
 | | HCV NS5B allosteric inhibitor complex | | Descriptor: | 4-METHYL-N-{(5E)-5-[(5-METHYL-2-FURYL)METHYLENE]-4-OXO-4,5-DIHYDRO-1,3-THIAZOL-2-YL}BENZENESULFONAMIDE, RNA-directed RNA polymerase (NS5B) (p68) | | Authors: | Yao, N. | | Deposit date: | 2006-08-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of a novel thiazolone scaffold as HCV NS5B polymerase allosteric inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HWI
 
 | | HCV NS5B allosteric inhibitor complex | | Descriptor: | (2S)-({(5Z)-5-[(5-ETHYL-2-FURYL)METHYLENE]-4-OXO-4,5-DIHYDRO-1,3-THIAZOL-2-YL}AMINO)(4-FLUOROPHENYL)ACETIC ACID, RNA-directed RNA polymerase (NS5B) (p68) | | Authors: | Yao, N. | | Deposit date: | 2006-08-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a novel thiazolone scaffold as HCV NS5B polymerase allosteric inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4NK3
 
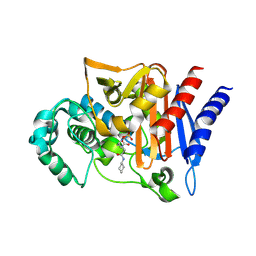 | | Amp-c beta-lactamase (pseudomonas aeruginosa) in complex with mk-7655 | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of MK-7655, a beta-lactamase inhibitor for combination with Primaxin().
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3F81
 
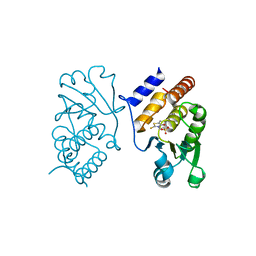 | | Interaction of VHR with SA3 | | Descriptor: | 2-[(5~{E})-5-[[3-[4-(2-fluoranylphenoxy)phenyl]-1-phenyl-pyrazol-4-yl]methylidene]-4-oxidanylidene-2-sulfanylidene-1,3-thiazolidin-3-yl]ethanesulfonic acid, Dual specificity protein phosphatase 3 | | Authors: | Wu, S, Mutelin, T, Tautz, L. | | Deposit date: | 2008-11-11 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multidentate small-molecule inhibitors of vaccinia H1-related (VHR) phosphatase decrease proliferation of cervix cancer cells.
J.Med.Chem., 52, 2009
|
|
3BYM
 
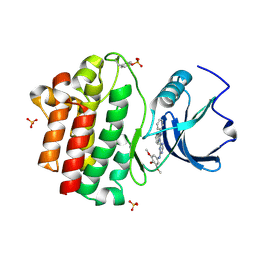 | | X-ray co-crystal structure aminobenzimidazole triazine 1 bound to Lck | | Descriptor: | N-phenyl-1-{4-[(3,4,5-trimethoxyphenyl)amino]-1,3,5-triazin-2-yl}-1H-benzimidazol-2-amine, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel 2-amino-6-phenyl-pyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-ones as potent and orally active inhibitors of lymphocyte specific kinase (Lck): synthesis, SAR, and in vivo anti-inflammatory activity.
J.Med.Chem., 51, 2008
|
|
3BYO
 
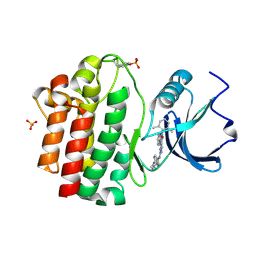 | | X-Ray co-crystal structure of 2-amino-6-phenylpyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-one 25 bound to Lck | | Descriptor: | 6-(2,6-dimethylphenyl)-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-one, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel 2-amino-6-phenyl-pyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-ones as potent and orally active inhibitors of lymphocyte specific kinase (Lck): synthesis, SAR, and in vivo anti-inflammatory activity.
J.Med.Chem., 51, 2008
|
|
2I1R
 
 | | Novel Thiazolones as HCV NS5B Polymerase Inhibitors: Further Designs, Synthesis, SAR and X-ray Complex Structure | | Descriptor: | (5Z)-5-[(5-ETHYL-2-FURYL)METHYLENE]-2-{[(S)-(4-FLUOROPHENYL)(1H-TETRAZOL-5-YL)METHYL]AMINO}-1,3-THIAZOL-4(5H)-ONE, RNA-directed RNA polymerase (NS5B) (P68) | | Authors: | Yao, N, Yan, S. | | Deposit date: | 2006-08-14 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel thiazolones as HCV NS5B polymerase allosteric inhibitors: Further designs, SAR, and X-ray complex structure.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1T8C
 
 | | Structure of the C-type lectin domain of CD23 | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor | | Authors: | Hibbert, R.G, Teriete, P, Grundy, G.J, Sutton, B.J, Gould, H.J, McDonnell, J.M. | | Deposit date: | 2004-05-12 | | Release date: | 2005-07-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of human CD23 and its interactions with IgE and CD21
J.Exp.Med., 202, 2005
|
|
2GQN
 
 | |
3L3L
 
 | | PARP complexed with A906894 | | Descriptor: | 3-oxo-2-piperidin-4-yl-2,3-dihydro-1H-isoindole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-12-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and SAR of substituted 3-oxoisoindoline-4-carboxamides as potent inhibitors of poly(ADP-ribose) polymerase (PARP) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3I28
 
 | | Crystal Structure of soluble epoxide Hydrolase | | Descriptor: | 4-cyano-N-{(3S)-3-(4-fluorophenyl)-3-[4-(methylsulfonyl)phenyl]propyl}benzamide, Epoxide hydrolase 2 | | Authors: | Farrow, N.A. | | Deposit date: | 2009-06-29 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based optimization of arylamides as inhibitors of soluble epoxide hydrolase.
J.Med.Chem., 52, 2009
|
|
3QC4
 
 | | PDK1 in complex with DFG-OUT inhibitor xxx | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Arndt, J.W. | | Deposit date: | 2011-01-15 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6KDZ
 
 | | Crystal structure of PDE10A in complex with a triazolopyrimidine inhibitor | | Descriptor: | 4-[2-(5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)ethyl]-3,7,8,10-tetrazatricyclo[7.4.0.0^{2,7}]trideca-1,3,5,8,10,12-hexaen-6-ol, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis, SAR study, and biological evaluation of novel 2,3-dihydro-1H-imidazo[1,2-a]benzimidazole derivatives as phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
