6DKS
 
 | | Structure of the Rbpj-SHARP-DNA Repressor Complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Kovall, R.A, VanderWielen, B.D, Yuan, Z. | | Deposit date: | 2018-05-30 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site.
Cell Rep, 26, 2019
|
|
1G5C
 
 | | CRYSTAL STRUCTURE OF THE 'CAB' TYPE BETA CLASS CARBONIC ANHYDRASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-CARBONIC ANHYDRASE, CALCIUM ION, ... | | Authors: | Strop, P, Smith, K.S, Iverson, T.M, Ferry, J.G, Rees, D.C. | | Deposit date: | 2000-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the "cab"-type beta class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum.
J.Biol.Chem., 276, 2001
|
|
6VPN
 
 | |
6L1L
 
 | |
6L1N
 
 | |
2MAN
 
 | | MANNOTRIOSE COMPLEX OF THERMOMONOSPORA FUSCA BETA-MANNANASE | | Descriptor: | PROTEIN (BETA-MANNANASE), beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Hilge, M, Gloor, S.M, Piontek, K. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution native and complex structures of thermostable beta-mannanase from Thermomonospora fusca - substrate specificity in glycosyl hydrolase family 5.
Structure, 6, 1998
|
|
9AZB
 
 | | Crystal structure of LolTv5 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-11 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
9AZA
 
 | | Crystal structure of LolTv4 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
5IPR
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 3 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
3T32
 
 | | Crystal structure of a putative C-S lyase from Bacillus anthracis | | Descriptor: | Aminotransferase, class I/II | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Peterson, S.N, Porebski, P, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-24 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative C-S lyase from Bacillus anthracis
TO BE PUBLISHED
|
|
5IPQ
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 2 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
1X1Z
 
 | | Orotidine 5'-monophosphate decarboxylase (odcase) complexed with BMP (produced from 6-cyanoump) | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fujihashi, M, Bello, A.M, Poduch, E, Wei, L, Annedi, S.C, Pai, E.F, Kotra, L.P. | | Deposit date: | 2005-04-15 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | An unprecedented twist to ODCase catalytic activity
J.Am.Chem.Soc., 127, 2005
|
|
1G7S
 
 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1LOS
 
 | |
4Y4N
 
 | | Thiazole synthase Thi4 from Methanococcus igneus | | Descriptor: | 2-[(E)-[(4R)-5-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-3-oxidanylidene-pentan-2-ylidene]amino]ethanoic acid, FE (II) ION, Putative ribose 1,5-bisphosphate isomerase | | Authors: | Zhang, X, Ealick, S.E. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
1G7T
 
 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B COMPLEXED WITH GDPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
7QX0
 
 | | Transaminase Structure of Plurienzyme (Tr2E2) in complex with PLP | | Descriptor: | Aminotransferase TR2, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-26 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
1LOR
 
 | | crystal structure of orotidine 5'-monophosphate complexed with BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, orotidine monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
7QYF
 
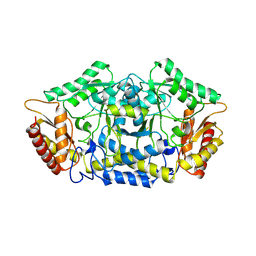 | | Structure of the transaminase PluriZyme variant (TR2E2) | | Descriptor: | Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-28 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
7QYG
 
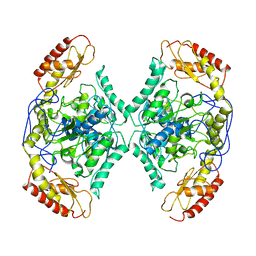 | | Structure of the transaminase TR2 | | Descriptor: | Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
5IPV
 
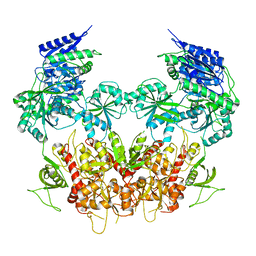 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 1 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, McHaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.25 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
1LOL
 
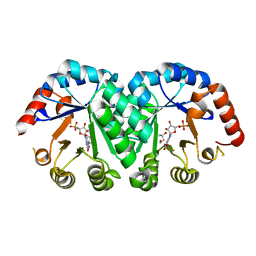 | | Crystal structure of orotidine monophosphate decarboxylase complex with XMP | | Descriptor: | 1,3-BUTANEDIOL, XANTHOSINE-5'-MONOPHOSPHATE, orotidine 5'-monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
1IX5
 
 | | Solution structure of the Methanococcus thermolithotrophicus FKBP | | Descriptor: | FKBP | | Authors: | Suzuki, R, Nagata, K, Kawakami, M, Nemoto, N, Furutani, M, Adachi, K, Maruyama, T, Tanokura, M. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Solution Structure of an Archaeal FKBP with a Dual Function of Peptidyl Prolyl cis-trans Isomerase and Chaperone-like Activities
J.MOL.BIOL., 328, 2003
|
|
4QE7
 
 | | Open MthK pore structure soaked in 10 mM Ba2+/100 mM Na+ | | Descriptor: | BARIUM ION, Calcium-gated potassium channel MthK | | Authors: | Guo, R, Zeng, W, Cui, H, Chen, L, Ye, S. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ionic interactions of Ba2+ blockades in the MthK K+ channel
J.Gen.Physiol., 144, 2014
|
|
2E6Y
 
 | | Covalent complex of orotidine 5'-monophosphate decarboxylase (ODCase) with 6-Iodo-UMP | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fujihashi, M, Bello, A.M, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Potent, Covalent Inhibitor of Orotidine 5'-Monophosphate Decarboxylase with Antimalarial Activity.
J.Med.Chem., 50, 2007
|
|
