4ZXL
 
 | | CpOGA D298N in complex with Drosophila HCF -derived Thr-O-GlcNAc peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, NAG-PRO-SER-THR-ALA-Thr-O-GlcNAc containing peptide from drosophila HCF, ... | | Authors: | Selvan, N, van Aalten, D.M.F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A mutant O-GlcNAcase as a probe to reveal global dynamics of protein O-GlcNAcylation during Drosophila embryonic development.
Biochem.J., 470, 2015
|
|
4ZYM
 
 | |
7N8Q
 
 | |
6VTU
 
 | | DH717.1 Fab monomer in complex with man9 glycan | | Descriptor: | DH717.1 heavy chain, DH717.1 light chain, GLYCEROL, ... | | Authors: | Fera, D, Bronkema, N. | | Deposit date: | 2020-02-13 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
5DT1
 
 | |
6CBJ
 
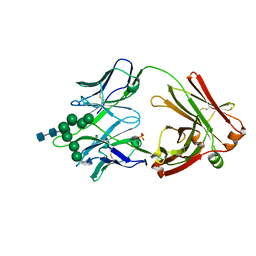 | | Crystal Structure of DH270.3 Fab in complex with Man9 | | Descriptor: | DH270.3 Fab heavy chain, DH270.3 Fab light chain, PHOSPHATE ION, ... | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | HIV envelope V3 region mimic embodies key features of a broadly neutralizing antibody lineage epitope.
Nat Commun, 9, 2018
|
|
6PM9
 
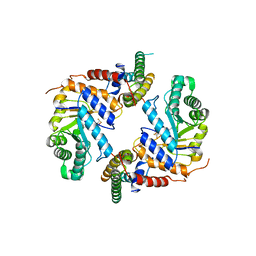 | | Crystal structure of the core catalytic domain of human O-GlcNAcase bound to MK-8719 | | Descriptor: | (3aR,5S,6S,7R,7aR)-5-(difluoromethyl)-2-(ethylamino)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Selnick, H.G, Duffy, J.L, McEachern, E.J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies.
J.Med.Chem., 62, 2019
|
|
5I5X
 
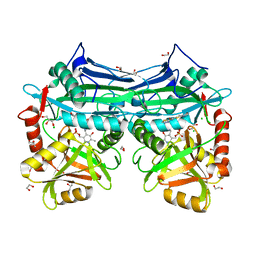 | | X-RAY CRYSTAL STRUCTURE AT 1.65A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A THIAZOLE COMPOUND AND PMP COFACTOR. | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 5-methyl-4-oxo-N-(1,3,4-thiadiazol-2-yl)-3,4-dihydrothieno[2,3-d]pyrimidine-6-carboxamide, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening.
J.Med.Chem., 59, 2016
|
|
6FUI
 
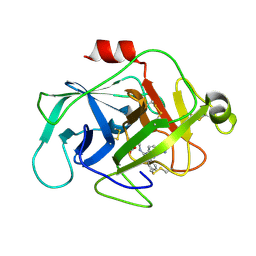 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | Descriptor: | (1~{R},2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]cyclohexane-1-carboxylic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6D20
 
 | | Crystal structure of Tyrosine-protein kinase receptor in complex with 5-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4(3H)-one and 5-{[2,4-dichloro-5-(pyridin-2-yl)benzene-1-carbonyl]amino}-N-(2-hydroxy-2-methylpropyl)-1-phenyl-1H-pyrazole-3-carboxamide Inhibitors | | Descriptor: | 5-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4(3H)-one, 5-{[2,4-dichloro-5-(pyridin-2-yl)benzene-1-carbonyl]amino}-N-(2-hydroxy-2-methylpropyl)-1-phenyl-1H-pyrazole-3-carboxamide, High affinity nerve growth factor receptor | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Allosteric, Potent, Subtype Selective, and Peripherally Restricted TrkA Kinase Inhibitors.
J. Med. Chem., 62, 2019
|
|
6D1W
 
 | | human PKD2 F604P mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Zheng, W, Yang, X, Bulkley, D, Chen, X.Z, Cao, E. | | Deposit date: | 2018-04-12 | | Release date: | 2018-06-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Hydrophobic pore gates regulate ion permeation in polycystic kidney disease 2 and 2L1 channels.
Nat Commun, 9, 2018
|
|
5E0A
 
 | | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A
To Be Published
|
|
7KV9
 
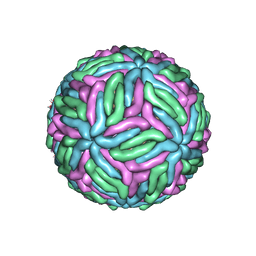 | | Chimeric flavivirus between Binjari virus and West Nile (Kunjin) virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Matrix protein M, envelope protein E | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-11-27 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
7KVA
 
 | | Structure of West Nile virus (Kunjin) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Matrix protein M | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-11-27 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
6D1Z
 
 | | Crystal structure of Tyrosine-protein kinase receptor in complex with 5-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4(3H)-one Inhibitor | | Descriptor: | 5-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4(3H)-one, 5-{[5-(6-aminopyridin-2-yl)-2-chlorobenzene-1-carbonyl]amino}-1-phenyl-1H-pyrazole-3-carboxamide, GLYCEROL, ... | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Allosteric, Potent, Subtype Selective, and Peripherally Restricted TrkA Kinase Inhibitors.
J. Med. Chem., 62, 2019
|
|
8CMO
 
 | | Cryo-EM structure of the Photosystem I - LHCI supercomplex from Coelastrella sp. | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Fadeeva, M, Klaiman, D, Nelson, N. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of the Photosystem I - LHCI supercomplex from Coelastrella sp.
To Be Published
|
|
6PQI
 
 | |
3HS5
 
 | | X-ray crystal structure of arachidonic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
3HS6
 
 | | X-ray crystal structure of eicosapentaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
4PDD
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM POLAROMONAS SP JS666 (Bpro_0088, TARGET EFI-510167) BOUND TO D-ERYTHRONATE | | Descriptor: | (2R,3R)-2,3,4-trihydroxybutanoic acid, FORMIC ACID, MALONIC ACID, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4XDA
 
 | | Vibrio cholerae O395 Ribokinase complexed with Ribose, ADP and Sodium ion. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribokinase, SODIUM ION, ... | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Apo and Ligand Bound Vibrio cholerae Ribokinase (Vc-RK): Role of Monovalent Cation Induced Activation and Structural Flexibility in Sugar Phosphorylation
Adv.Exp.Med.Biol., 842, 2015
|
|
6SND
 
 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
6G7N
 
 | | Trichodesmium Tery_3377 (IdiA) (FutA) with iron and alanine ligand. | | Descriptor: | D-ALANINE, D-GLUTAMIC ACID, Extracellular solute-binding protein, ... | | Authors: | Machelett, M.M, Tews, I. | | Deposit date: | 2018-04-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and functional characterization of IdiA/FutA (Tery_3377), an iron-binding protein from the ocean diazotrophTrichodesmium erythraeum.
J. Biol. Chem., 293, 2018
|
|
5LFA
 
 | | Crystal structure of iron-sulfur cluster containing bacterial (6-4) photolyase PhrB - Y424F mutant with impaired DNA repair activity | | Descriptor: | (6-4) photolyase, 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kwiatkowski, D, Zhang, F, Krauss, N, Lamparter, T, Scheerer, P. | | Deposit date: | 2016-06-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
5TD4
 
 | |
