4OK6
 
 | |
6HTR
 
 | | Yeast 20S proteasome with human beta2c (S171G) in complex with 13 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, MAGNESIUM ION, PROTEASOME SUBUNIT ALPHA TYPE-1, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HW3
 
 | | Yeast 20S proteasome in complex with 13 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6FB0
 
 | |
5PHL
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D in complex with N09506a | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-1H-indole-2,3-dione, Lysine-specific demethylase 4D, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.141 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5EJV
 
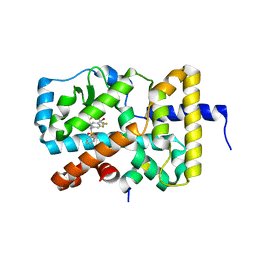 | | RORy in complex with T090131718 and Coactivator peptide EBI96 | | Descriptor: | EBI96 Coactivator Peptide, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, Nuclear receptor ROR-gamma | | Authors: | Marcotte, D.M. | | Deposit date: | 2015-11-02 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of biaryls as ROR gamma inverse agonists by using structure-based design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4YMB
 
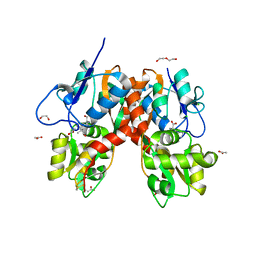 | | Structure of the ligand-binding domain of GluK1 in complex with the antagonist CNG10111 | | Descriptor: | (3R,4S)-3-(3-carboxyphenyl)-4-propyl-L-proline, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Moller, C, Tapken, D, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2015-03-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-Activity Relationship Study of Ionotropic Glutamate Receptor Antagonist (2S,3R)-3-(3-Carboxyphenyl)pyrrolidine-2-carboxylic Acid.
J.Med.Chem., 58, 2015
|
|
4B34
 
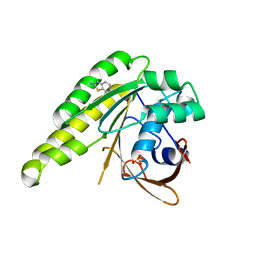 | | Humanised monomeric RadA in complex with 2-amino benzothiazole | | Descriptor: | 1,3-benzothiazol-2-amine, DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-20 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
6SY7
 
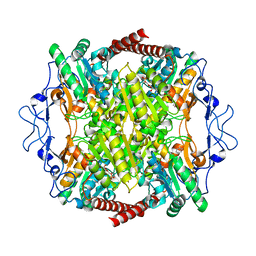 | | Structure of Trypanosome Brucei Phosphofructokinase in complex with AMP. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent 6-phosphofructokinase, BENZENE, ... | | Authors: | McNae, I.W, Vasquez-Valdivieso, M.G, Walkinshaw, M.D. | | Deposit date: | 2019-09-27 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Kinetic and structural studies of Trypanosoma and Leishmania phosphofructokinases show evolutionary divergence and identify AMP as a switch regulating glycolysis versus gluconeogenesis.
Febs J., 287, 2020
|
|
2WP7
 
 | | Crystal structure of deSUMOylase(DUF862) | | Descriptor: | PPPDE PEPTIDASE DOMAIN-CONTAINING PROTEIN 2 | | Authors: | Kim, J.H, Woo, J.S, Oh, B.H. | | Deposit date: | 2009-08-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Desi-1, a Novel Desumoylase Belonging to a Putative Isopeptidase Superfamily.
Proteins, 80, 2012
|
|
4OK3
 
 | |
3NC9
 
 | |
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
7EC8
 
 | |
3UMJ
 
 | | Crystal Structure of D311E Lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ruslan, R, Rahman, R.N.Z.R.A, Leow, T.C, Ali, M.S.M, Basri, M, Salleh, A.B. | | Deposit date: | 2011-11-13 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Thermal Stability via Outer-Loop Ion Pair Interaction of Mutated T1 Lipase from Geobacillus zalihae Strain T1
Int J Mol Sci, 13, 2012
|
|
2X0V
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 4-(trifluoromethyl)benzene-1,2-diamine | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Basse, N, Kaar, J.L, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
6SFK
 
 | | Crystal structure of p38 alpha in complex with compound 81 (MCP42) | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
5EQZ
 
 | |
4PLW
 
 | | Crystal structure of ancestral apicomplexan malate dehydrogenase with lactate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PHOSPHATE ION, malate dehydrogenase | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-19 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
4PLC
 
 | | Crystal structure of ancestral apicomplexan lactate dehydrogenase with malate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID, ... | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
6SOD
 
 | | Fragment N14056a in complex with MAP kinase p38-alpha | | Descriptor: | 1-[[(3~{S})-1,4-dioxaspiro[4.5]decan-3-yl]methyl]piperidine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nichols, C.E, De Nicola, G.F. | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
9CF6
 
 | |
6SR4
 
 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 112 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
6B8J
 
 | |
3F5L
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-04 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
