3EB3
 
 | | Voltage-dependent K+ channel beta subunit (W121A) in complex with cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel subunit beta-2 | | Authors: | Pan, Y, Weng, J, Kabaleeswaran, V, Li, H, Cao, Y, Bhosle, R.C, Zhou, M. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cortisone dissociates the Shaker family K+ channels from their beta subunits.
Nat.Chem.Biol., 4, 2008
|
|
6KXX
 
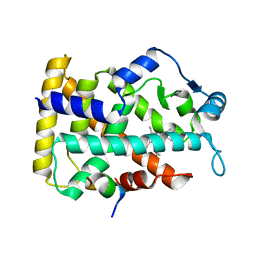 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist (compound A) | | Descriptor: | 1-(4-chlorophenyl)-6-methyl-3-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, PGC1alpha, Peroxisome proliferator-activated receptor alpha | | Authors: | Yoshida, T, Tachibana, K, Oki, H, Doi, M, Fukuda, S, Yuzuriha, T, Tabata, R, Ishimoto, K, Kawahara, K, Ohkubo, T, Miyachi, H, Doi, T. | | Deposit date: | 2019-09-14 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for PPAR alpha Activation by 1H-pyrazolo-[3,4-b]pyridine Derivatives.
Sci Rep, 10, 2020
|
|
5UI1
 
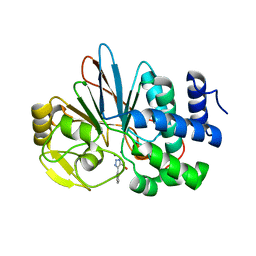 | | Crystal Structure of Human Protein Phosphatase 5C (PP5C) in complex with a triazole inhibitor | | Descriptor: | 5-phenyl-1H-1,2,3-triazole-4-carboxylic acid, MANGANESE (II) ION, Serine/threonine-protein phosphatase 5 | | Authors: | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Banerjee, S, Honkanen, R.E. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure Human PP5C in Complex with an Inhibitor
To Be Published
|
|
2BCQ
 
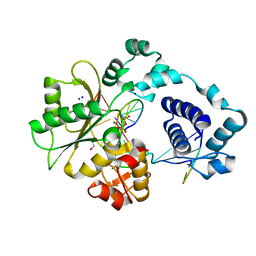 | | DNA polymerase lambda in complex with a DNA duplex containing an unpaired Dtmp | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*TP*TP*AP*CP*TP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2005-10-19 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase
Cell(Cambridge,Mass.), 124, 2006
|
|
7QTJ
 
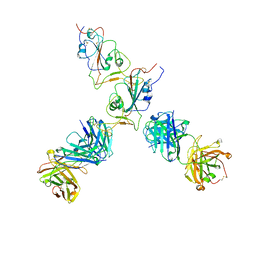 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, P2G3 Light Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
4M6Q
 
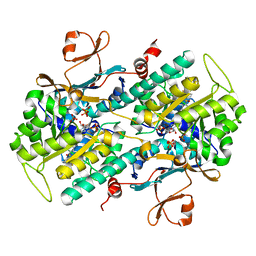 | | Identification of Amides Derived From 1H-Pyrazolo[3,4-b]pyridine-5-carboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-(5-O-phosphono-beta-D-ribofuranosyl)-N-(4-{[3-(trifluoromethyl)phenyl]sulfonyl}benzyl)-1H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-08-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R8W
 
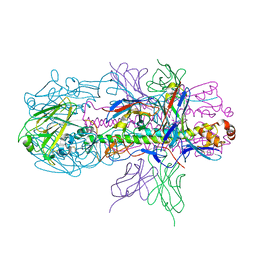 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a neutralizing antibody CT149 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of neutralizing antibody CT149, Hemagglutinin, ... | | Authors: | Wu, Y, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus
Nat Commun, 6, 2015
|
|
4R9M
 
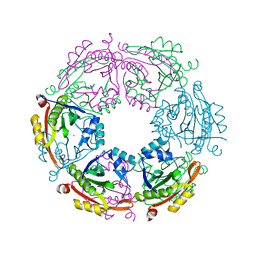 | | Crystal structure of spermidine N-acetyltransferase from Escherichia coli | | Descriptor: | MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8AI7
 
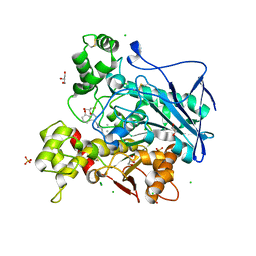 | | Structure of carbamoylated human butyrylcholinesterase upon reaction with 3-(((2-cycloheptylethyl)(methyl)amino)methyl)-1H-indol-7-yl N,N-dimethylcarbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[2-cycloheptylethyl(methyl)amino]methyl]-1~{H}-indol-7-ol, ... | | Authors: | Brazzolotto, X, Meden, A, Knez, D, Gobec, S, Nachon, F. | | Deposit date: | 2022-07-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Pseudo-irreversible butyrylcholinesterase inhibitors: Structure-activity relationships, computational and crystallographic study of the N-dialkyl O-arylcarbamate warhead.
Eur.J.Med.Chem., 247, 2023
|
|
4YQI
 
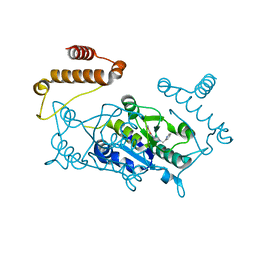 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | (4-amino-1,2,5-oxadiazol-3-yl)[4-(pyridin-2-ylmethyl)piperazin-1-yl]methanone, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Madauss, K.P, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
6KSN
 
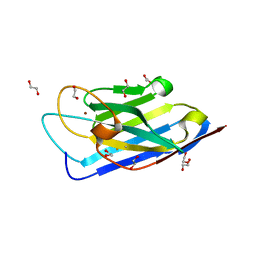 | | Structure of a Zn-bound camelid single domain antibody | | Descriptor: | 1,2-ETHANEDIOL, ICab3, SODIUM ION, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2019-08-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Isolation and structural characterization of a Zn2+-bound single-domain antibody against NorC, a putative multidrug efflux transporter in bacteria.
J.Biol.Chem., 295, 2020
|
|
4YQS
 
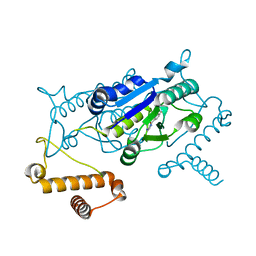 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | (5-amino-1H-1,2,4-triazol-1-yl)(4-methoxyphenyl)methanone, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
6Y0F
 
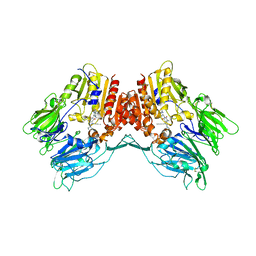 | | Structure of human FAPalpha in complex with linagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8-[(3R)-3-Aminopiperidin-1-yl]-7-but-2-yn-1-yl-3-methyl-1-[(4-methylquinazolin-2-yl)methyl]-3,7-dihydro-1H-purine-2,6-d ione, ... | | Authors: | Nar, H, Schnapp, G, Schreiner, P. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Structure of human FAPalpha in complex with linagliptin
To Be Published
|
|
4YRE
 
 | |
6T6G
 
 | | Bacteroides salyersiae GH164 beta-mannosidase in complex with noeuromycin | | Descriptor: | (2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Armstrong, Z, Davies, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure and function ofBs164 beta-mannosidase fromBacteroides salyersiaethe founding member of glycoside hydrolase family GH164.
J.Biol.Chem., 295, 2020
|
|
2WKI
 
 | | Crystal structure of the OXA-10 K70C mutant at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GLYCEROL, ... | | Authors: | Vercheval, L, Bauvois, C, Kerff, F, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
8CG9
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with a cyclic N-hydroxyurea inhibitor | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, 1-[[(3~{R})-2,3-dihydro-1,4-benzodioxin-3-yl]methyl]-3-oxidanyl-thieno[3,2-d]pyrimidine-2,4-dione, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
4YFI
 
 | | TNNI3K complexed with inhibitor 1 | | Descriptor: | N-methyl-3-(9H-purin-6-ylamino)benzenesulfonamide, Serine/threonine-protein kinase TNNI3K | | Authors: | Shewchuk, L.M, Wang, L, Lawhorn, B.G. | | Deposit date: | 2015-02-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of Purines and 7-Deazapurines as Potent and Selective Type I Inhibitors of Troponin I-Interacting Kinase (TNNI3K).
J.Med.Chem., 58, 2015
|
|
4MC8
 
 | | Hedycaryol synthase in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative sesquiterpene cyclase | | Authors: | Baer, P, Rabe, P, Cirton, C, Oliveira Mann, C, Kaufmann, N, Groll, M, Dickschat, J. | | Deposit date: | 2013-08-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hedycaryol synthase in complex with nerolidol reveals terpene cyclase mechanism.
Chembiochem, 15, 2014
|
|
7MGY
 
 | | Sco GlgEI-V279S in complex with 4-alpha-glucoside of valienamine | | Descriptor: | (1R,4S,5S,6R)-4-amino-5,6-dihydroxy-2-(hydroxymethyl)cyclohex-2-en-1-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereoselective synthesis of a 4-⍺-glucoside of valienamine and its X-ray structure in complex with Streptomyces coelicolor GlgE1-V279S.
Sci Rep, 11, 2021
|
|
7MEL
 
 | | Sco GlgEI-V279S in complex with 4-alpha-glucoside of validamine | | Descriptor: | (1R,2R,3S,4S,6R)-4-amino-2,3-dihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-04-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stereoselective synthesis of a 4-alpha-glucoside of valienamine and its X-ray structure in complex with Streptomyces coelicolor GlgE1-V279S.
Sci Rep, 11, 2021
|
|
7HOO
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with ASAP-0014643-001 | | Descriptor: | (2P)-2-(5-methoxypyridin-3-yl)-N-(1-methyl-1H-pyrazol-4-yl)benzamide, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Lee, A, Kenton, N, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2024-11-07 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2WNC
 
 | | Crystal structure of Aplysia ACHBP in complex with tropisetron | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-INDOLE-3-CARBOXYLATE, Soluble acetylcholine receptor | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
EMBO J., 28, 2009
|
|
9CV2
 
 | |
