3W6X
 
 | | Yeast N-acetyltransferase Mpr1 in complex with CHOP | | Descriptor: | (4S)-4-hydroxy-L-proline, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1U8B
 
 | | Crystal structure of the methylated N-ADA/DNA complex | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*TP*T)-3', 5'-D(P*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*AP*T)-3', ... | | Authors: | He, C, Hus, J.-C, Sun, L.J, Zhou, P, Norman, D.P.G, Dotsch, V, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2004-08-05 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada.
Mol.Cell, 20, 2005
|
|
3WCE
 
 | | The complex structure of TcSQS with ligand, ER119884 | | Descriptor: | (3R)-3-{[2-benzyl-6-(3-methoxypropoxy)pyridin-3-yl]ethynyl}-1-azabicyclo[2.2.2]octan-3-ol, Farnesyltransferase, putative | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3WCM
 
 | | The complex structure of HsSQS wtih ligand, ER119884 | | Descriptor: | (3R)-3-{[2-benzyl-6-(3-methoxypropoxy)pyridin-3-yl]ethynyl}-1-azabicyclo[2.2.2]octan-3-ol, Squalene synthase | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-28 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3A0W
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) for MAD phasing (nucleotide free form 2, orthorombic) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(ETHYLMERCURI-THIO)-BENZOIC ACID, ETHYL MERCURY ION, ... | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
1TVV
 
 | |
3WCD
 
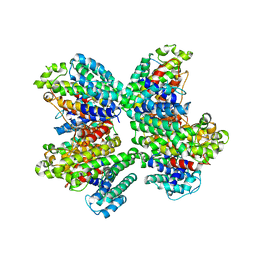 | | The complex structure of HsSQS wtih ligand, WC-9 | | Descriptor: | 2-(4-phenoxyphenoxy)ethyl thiocyanate, Squalene synthase | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
1TVW
 
 | |
2NPO
 
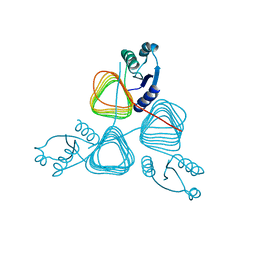 | | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Acetyltransferase | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
1TSZ
 
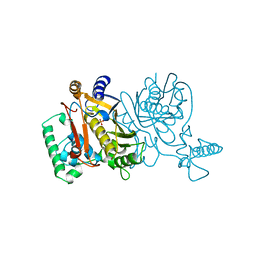 | | THYMIDYLATE SYNTHASE R179K MUTANT | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1TSW
 
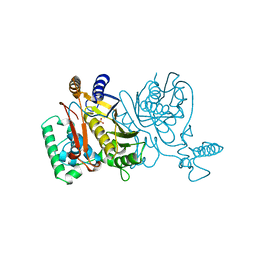 | | THYMIDYLATE SYNTHASE R179A MUTANT | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
5UJS
 
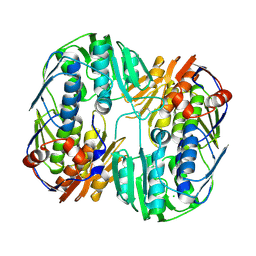 | | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni.
To Be Published
|
|
1TSL
 
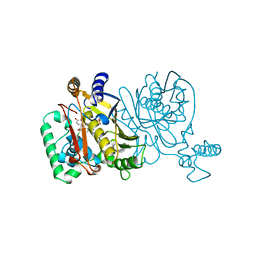 | |
1TSM
 
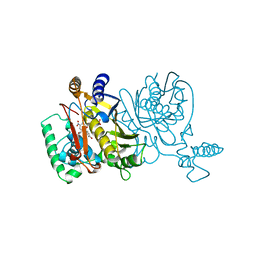 | |
3A25
 
 | | Crystal structure of P. horikoshii TYW2 in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, Uncharacterized protein PH0793 | | Authors: | Umitsu, M, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1L0C
 
 | | Investigation of the Roles of Catalytic Residues in Serotonin N-Acetyltransferase | | Descriptor: | COA-S-ACETYL TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Scheibner, K.A, De Angelis, J, Burley, S.K, Cole, P.A. | | Deposit date: | 2002-02-08 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of the roles of catalytic residues in serotonin N-acetyltransferase.
J.Biol.Chem., 277, 2002
|
|
3WCJ
 
 | | The complex structure of HsSQS wtih ligand,E5700 | | Descriptor: | (3R)-3-({2-benzyl-6-[(3R,4S)-3-hydroxy-4-methoxypyrrolidin-1-yl]pyridin-3-yl}ethynyl)-1-azabicyclo[2.2.2]octan-3-ol, Squalene synthase | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
1UBV
 
 | |
3WQM
 
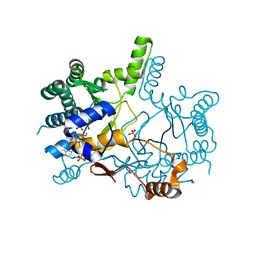 | | Crystal structure of Rv3378c with inhibitor BPH-629 | | Descriptor: | Diterpene synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
4L8A
 
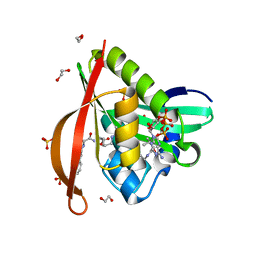 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in ternary complex with N-Phenylacetyl-Gly-AcLys and CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, N-(phenylacetyl)glycyl-N~6~-acetyl-L-lysine, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-06-16 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
1L9B
 
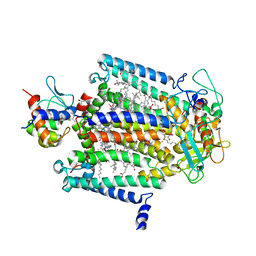 | | X-Ray Structure of the Cytochrome-c(2)-Photosynthetic Reaction Center Electron Transfer Complex from Rhodobacter sphaeroides in Type II Co-Crystals | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CHLORIDE ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Okamura, M.Y, Yeh, A.P, Rees, D.C, Feher, G. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure determination of the cytochrome c2: reaction center electron transfer complex from Rhodobacter sphaeroides.
J.Mol.Biol., 319, 2002
|
|
1G0W
 
 | |
1UAE
 
 | | STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, [(1R)-1-hydroxypropyl]phosphonic acid | | Authors: | Skarzynski, T. | | Deposit date: | 1996-09-30 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Structure, 4, 1996
|
|
1UBW
 
 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | FARNESYL DIPHOSPHATE SYNTHASE, GERANYL DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1UBX
 
 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | FARNESYL DIPHOSPHATE, FARNESYL DIPHOSPHATE SYNTHASE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
