7RD2
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 2 | | Descriptor: | (2R,3R,4R,5S)-1-{[4-({4-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-2-nitroanilino}methyl)phenyl]methyl}-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-09 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
5ELZ
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with a pantothenate analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, N-(1,3-benzodioxol-5-ylmethyl)-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Mottaghi, K, Hughes, S.J, Tempel, W, Hong, B, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-05 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent Pantothenamide Inhibitors of Staphylococcus aureus Pantothenate Kinase through a Minimal SAR Study: Inhibition Is Due to Trapping of the Product.
Acs Infect Dis., 2, 2016
|
|
6DNS
 
 | | Endo-fucoidan hydrolase MfFcnA9 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
4YOM
 
 | | Structure of SAD kinase | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase BRSK2 | | Authors: | Wu, J.X, Wang, J, Chen, L, Wang, Z.X, Wu, J.W. | | Deposit date: | 2015-03-12 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insight into the mechanism of synergistic autoinhibition of SAD kinases
Nat Commun, 6, 2015
|
|
4YQL
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | 4-amino-N-[(1S,2R)-2-(4-sulfamoylphenyl)cyclopropyl]-1,2,5-oxadiazole-3-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Madauss, K.P. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
4YQQ
 
 | |
3FMW
 
 | | The crystal structure of MtmOIV, a Baeyer-Villiger monooxygenase from the mithramycin biosynthetic pathway in Streptomyces argillaceus. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase | | Authors: | Noinaj, N, Beam, M.P, Wang, C, Rohr, J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of Baeyer-Villiger monooxygenase MtmOIV, the key enzyme of the mithramycin biosynthetic pathway .
Biochemistry, 48, 2009
|
|
7M3Z
 
 | | Structure of TIM-3 in complex with N-(4-(8-chloro-2-mehtyl-5-oxo-5,6-dihydro-[1,2,4]triazolo[1,5-c]quinazolin-9-yl)-3-methylphenyl)methanesulfonamdide (compound 35) | | Descriptor: | CALCIUM ION, Hepatitis A virus cellular receptor 2, N-{4-[(4S,10aP)-8-chloro-2-methyl-5-oxo-5,6-dihydro[1,2,4]triazolo[1,5-c]quinazolin-9-yl]-3-methylphenyl}methanesulfonamide | | Authors: | Rietz, T.A. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment-Based Discovery of Small Molecules Bound to T-Cell Immunoglobulin and Mucin Domain-Containing Molecule 3 (TIM-3).
J.Med.Chem., 64, 2021
|
|
4EHC
 
 | |
7M3Y
 
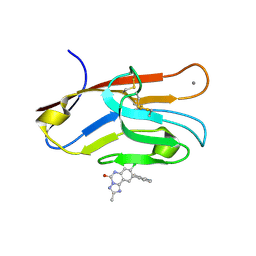 | | Structure of TIM-3 in complex with 8-chloro-2-methyl-9-(3-mehtylpyridin-4-yl)-[1,2,4]triazolo[1,5-c]quinazolin-5(6H)-one (compound 22) | | Descriptor: | (4R,10aP)-8-chloro-2-methyl-9-(3-methylpyridin-4-yl)[1,2,4]triazolo[1,5-c]quinazolin-5(6H)-one, CALCIUM ION, Hepatitis A virus cellular receptor 2 | | Authors: | Rietz, T.A. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment-Based Discovery of Small Molecules Bound to T-Cell Immunoglobulin and Mucin Domain-Containing Molecule 3 (TIM-3).
J.Med.Chem., 64, 2021
|
|
8GFW
 
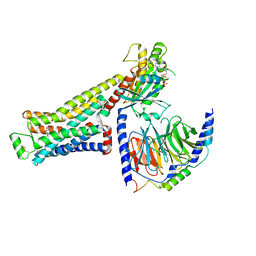 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #2 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GFX
 
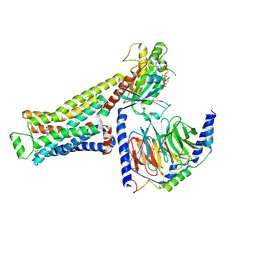 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #3 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
7MIP
 
 | | Mouse CTPS1 bound to inhibitor R80 | | Descriptor: | CTP synthase 1, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8GFY
 
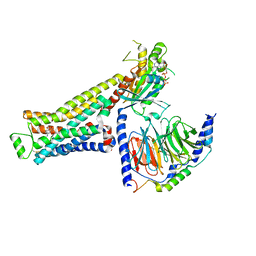 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #4 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
4Y9E
 
 | | Crystal structure of V30M mutated transthyretin in complex with gamma-mangostin | | Descriptor: | 1,3,6,7-tetrahydroxy-2,8-bis(3-methylbut-2-en-1-yl)-9H-xanthen-9-one, CHLORIDE ION, Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2015-02-17 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of gamma-Mangostin as an Amyloidogenesis Inhibitor
Sci Rep, 5, 2015
|
|
8V51
 
 | | Crystal structure of a HLA-B*35:01-NP10 with D1 TCR | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, D1 TCR alpha chain, ... | | Authors: | Littler, D.R, Rossjohn, J, Gras, S. | | Deposit date: | 2023-11-30 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.10000038 Å) | | Cite: | Molecular determinants of cross-strain influenza A virus recognition by alpha beta T cell receptors.
Sci Immunol, 10, 2025
|
|
8GFZ
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #5 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GG0
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #6 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
2X18
 
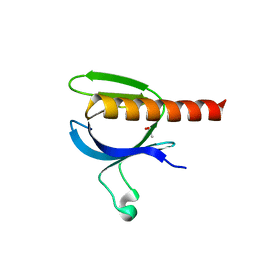 | | The crystal structure of the PH domain of human AKT3 protein kinase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, RAC-GAMMA SERINE/THREONINE-PROTEIN KINASE | | Authors: | Vollmar, M, Wang, J, Zhang, Y, Elkins, J.M, Burgess-Brown, N, Chaikuad, A, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2009-12-22 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The Crystal Structure of the Ph Domain of Human Akt3 Protein Kinase
To be Published
|
|
6SRD
 
 | |
3VP6
 
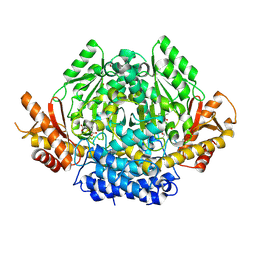 | | Structural characterization of Glutamic Acid Decarboxylase; insights into the mechanism of autoinactivation | | Descriptor: | 4-oxo-4H-pyran-2,6-dicarboxylic acid, GLYCEROL, Glutamate decarboxylase 1 | | Authors: | Langendorf, C.G, Tuck, K.L, Key, T.L.G, Rosado, C.J, Wong, A.S.M, Fenalti, G, Buckle, A.M, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the mechanism through which human glutamic acid decarboxylase auto-activates
Biosci.Rep., 33, 2013
|
|
8GG1
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #7 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GG2
 
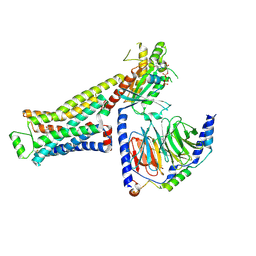 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #8 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
7MSD
 
 | | Structure of EED bound to EEDi-6068 | | Descriptor: | (9aP,12aR)-4-(2,2-difluoropropyl)-12-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,8,11,12a-pentaazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, FORMIC ACID, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
8VCK
 
 | | Galactose-binding lectin from Mytilus californianus, Isoform 1 (rMcL-1) | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Hernandez-Santoyo, A, Loera-Rubalcava, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A mytilectin from Mytilus californianus: Study of its unique galactoside interactions, oligomerization patterns, and antifungal activity.
Int.J.Biol.Macromol., 308, 2025
|
|
