7A0X
 
 | | Structure of dimeric sodium proton antiporter NhaA, at pH 6.0, crystallized with chimeric Fab antibodies | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, ... | | Authors: | Fippel, A, Lentes, C.J, Mir, S.H, Wirth, C, Hunte, C. | | Deposit date: | 2020-08-11 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Molecular determinants for substrate uptake in electrogenic sodium/proton antiporters
To Be Published
|
|
7V28
 
 | |
5VZ0
 
 | | Crystal structure of Lactococcus lactis pyruvate carboxylase G746A mutant in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5BXR
 
 | | LNBase in complex with LNB-NHAcDNJ | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
6YB9
 
 | | Human octameric PAICS in complex with SAICAR, AMP-PNP, and magnesium | | Descriptor: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Skerlova, J, Unterlass, J, Gottmann, M, Homan, E, Helleday, T, Jemth, A.S, Stenmark, P. | | Deposit date: | 2020-03-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Crystal structures of human PAICS reveal substrate and product binding of an emerging cancer target.
J.Biol.Chem., 295, 2020
|
|
7A0Y
 
 | | Structure of dimeric sodium proton antiporter NhaA K300R variant, at pH 8.2, crystallized with chimeric Fab antibodies | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Fippel, A, Mir, S.H, Lentes, C.J, Wirth, C, Hunte, C. | | Deposit date: | 2020-08-11 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular determinants for substrate uptake in electrogenic sodium/proton antiporters
To Be Published
|
|
8YSW
 
 | | phosphinothricin dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, Glutamate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Xue, Y.P, Cheng, F, Zhou, S.P, Xu, J.M, Jin, L.Q, Ma, C.J, Zheng, Y.G. | | Deposit date: | 2024-03-23 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of phosphinothricin dehydrogenase with NAD at 2.6 Angstorms resolution
To Be Published
|
|
5GU3
 
 | | Crystal structure of Au(E).CL-apo-E45C/R52C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Observation of gold sub-nanocluster nucleation within a crystalline protein cage
Nat Commun, 8, 2017
|
|
8FKF
 
 | |
8FKD
 
 | |
4QGP
 
 | |
8FKG
 
 | |
8FKC
 
 | |
6EQ8
 
 | |
8FKE
 
 | |
6T03
 
 | | Crystal structure of YTHDC1 with fragment 16 (DHU_DC1_017) | | Descriptor: | 1,3-dihydroimidazole-2-thione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
8ZIM
 
 | | Crystal structure of the methyltransferase PsiM in complex with SAH and Baeocystin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Baeocystin, GLYCEROL, ... | | Authors: | Meng, C.Y, Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-05-14 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for psilocybin biosynthesis.
Nat Commun, 16, 2025
|
|
8ABU
 
 | | Crystal structure of NaLdpA mutant H97Q in complex with erythro-DGPD | | Descriptor: | (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SnoaL-like domain-containing protein | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6MXT
 
 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
9G0D
 
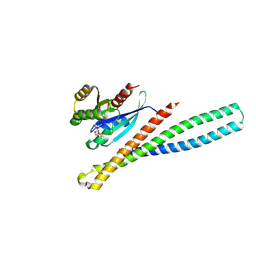 | | Structure of human Mical1 bMERB_V978A domain:Rab10 complex. | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-10, ... | | Authors: | Rai, A, Goody, R.S. | | Deposit date: | 2024-07-08 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and structural insights into the auto-inhibited state of Mical1 and its activation by Rab8
Elife, 2024
|
|
5M6D
 
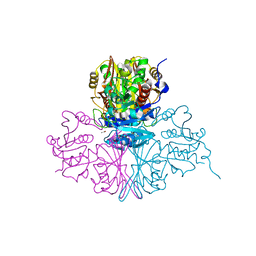 | | Streptococcus pneumoniae Glyceraldehyde-3-Phosphate Dehydrogenase (SpGAPDH) crystal structure | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Gaboriaud, C, Moreau, C.P, Di Guilmi, A.M. | | Deposit date: | 2016-10-25 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering Key Residues Involved in the Virulence-promoting Interactions between Streptococcus pneumoniae and Human Plasminogen.
J. Biol. Chem., 292, 2017
|
|
8ABW
 
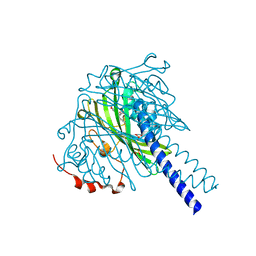 | | Crystal structure of SpLdpA in complex with threo-DGPD | | Descriptor: | (1R,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, (1S,2S)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SULFATE ION, ... | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5JRQ
 
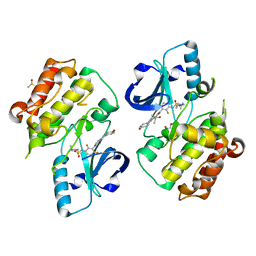 | | BRAFV600E Kinase Domain In Complex with Chemically Linked Vemurafenib Inhibitor VEM-6-VEM | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{2,4-difluoro-3-[5-(4-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]phenyl}propane-1-sulfonamide, ... | | Authors: | Grasso, M.J, Marmorstein, R. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Chemically Linked Vemurafenib Inhibitors Promote an Inactive BRAF(V600E) Conformation.
Acs Chem.Biol., 11, 2016
|
|
7S8W
 
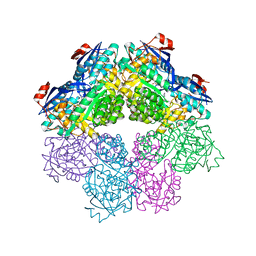 | | Amycolatopsis sp. T-1-60 N-succinylamino acid racemase/o-succinylbenzoate synthase R266Q mutant in complex with N-succinylphenylglycine | | Descriptor: | MAGNESIUM ION, N-succinyl-L-phenylglycine, N-succinylamino acid racemase/O-succinylbenzoate synthase, ... | | Authors: | Truong, D.P, Rousseau, S, Sacchettini, J.C, Glasner, M.E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Second-Shell Amino Acid R266 Helps Determine N -Succinylamino Acid Racemase Reaction Specificity in Promiscuous N -Succinylamino Acid Racemase/ o -Succinylbenzoate Synthase Enzymes.
Biochemistry, 60, 2021
|
|
5GIG
 
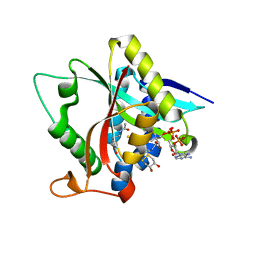 | | Crystal Structure of Drosophila melanogaster E47D Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Wu, C.Y, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-23 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Drosophila melanogaster E47D Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-dopamine
To Be Published
|
|
