8EAD
 
 | |
7N6Q
 
 | | Structure of PPPA bound human ACAT2 | | Descriptor: | (3S,4R,4aR,6S,6aS,12R,12aS,12bS)-4-[(acetyloxy)methyl]-12-hydroxy-4,6a,12b-trimethyl-11-oxo-9-(pyridin-3-yl)-1,3,4,4a,5,6,6a,12,12a,12b-decahydro-2H,11H-naphtho[2,1-b]pyrano[3,4-e]pyran-3,6-diyl diacetate, CHOLESTEROL, OLEIC ACID, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structure of PPPA bound human ACAT2
To Be Published
|
|
7EL4
 
 | | The crystal structure of p53p peptide fragment in complex with the N-terminal domain of MdmX | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Cellular tumor antigen p53,Protein Mdm4, MAGNESIUM ION | | Authors: | Cheng, X.Y, Zhang, B.L, Li, Z.C, Kuang, Z.K, Su, Z.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of the N-terminal domain of MdmX in complex with p53p peptide fragment
To Be Published
|
|
7M4Q
 
 | | Multidrug Efflux pump AdeJ | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7Q0I
 
 | | Crystal structure of the N-terminal domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
6ZB1
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH GSK620 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ~{N}5-cyclopropyl-~{N}3-methyl-2-oxidanylidene-1-(phenylmethyl)pyridine-3,5-dicarboxamide | | Authors: | Chung, C. | | Deposit date: | 2020-06-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6ZMB
 
 | | Structure of the native tRNA-Monooxygenase enzyme MiaE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
8ZBW
 
 | | Cryo-EM structure of formyl peptide receptor 2/C1R receptor in complex with Gi | | Descriptor: | C1R, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhou, Q, Lin, S, Li, G. | | Deposit date: | 2024-04-28 | | Release date: | 2025-09-10 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Oral FPR2/ALX modulators tune myeloid cell activity to ameliorate mucosal inflammation in inflammatory bowel disease.
Acta Pharmacol.Sin., 46, 2025
|
|
8DPD
 
 | |
4TMP
 
 | | Crystal structure of AF9 YEATS bound to H3K9ac peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-LYS-GLN-THR-ALA-ARG-ALY-SER-THR, Protein AF-9 | | Authors: | Li, H, Li, Y, Wang, H, Ren, Y. | | Deposit date: | 2014-06-02 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation.
Cell, 159, 2014
|
|
7UJR
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B | | Descriptor: | 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase type II subunit alpha, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Santos, N.J, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
6BEI
 
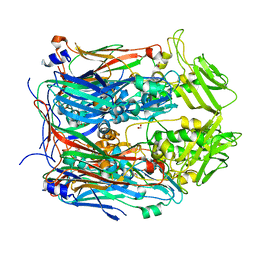 | | Crystal structure of VACV D13 in its apo (unbound) form | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Scaffold protein D13 | | Authors: | Garriga, D, Accurso, C, Coulibaly, F. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for the inhibition of poxvirus assembly by the antibiotic rifampicin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7PYB
 
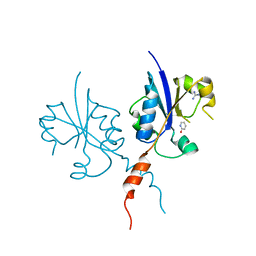 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 2-Hydroxypyridine | | Descriptor: | 1~{H}-pyridin-2-one, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-09 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
8VH8
 
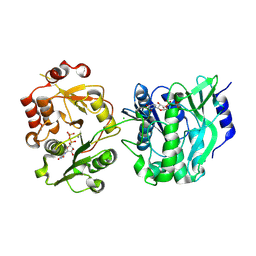 | | Crystal structure of heparosan synthase 2 from Pasteurella multocida at 2.85 A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pedersen, L.C, Liu, J, Stancanelli, E, krahn, J.M. | | Deposit date: | 2023-12-31 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Functional Analysis of Heparosan Synthase 2 from Pasteurella multocida (PmHS2) to Improve the Synthesis of Heparin.
Acs Catalysis, 14, 2024
|
|
8CAO
 
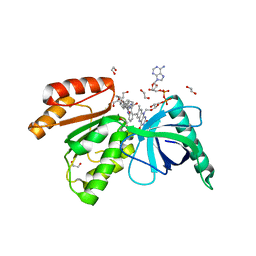 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with CA24 | | Descriptor: | 1,2-ETHANEDIOL, 8-[2-(4-cyclohexylphenyl)quinolin-4-yl]carbonyl-1,3,8-triazaspiro[4.5]decane-2,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
7WOI
 
 | | Structure of the shaft pilin Spa2 from Corynebacterium glutamicum | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Spa2 | | Authors: | Wu, Y.F, Wang, L.T, Huang, Y.Y, Zhong, C, Zhou, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Accelerating the design of pili-enabled living materials using an integrative technological workflow.
Nat.Chem.Biol., 2023
|
|
8RVB
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 8 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[2-(1~{H}-1,2,3-triazol-4-yl)ethylsulfanylmethyl]oxolane-3,4-diol, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Inhibition of the SARS-CoV-2 nsp16 by Substrate-Based Dual Site Inhibitors.
Chemmedchem, 19, 2024
|
|
6WJ5
 
 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
4TOB
 
 | | 1.95A resolution structure of BfrB (Q151L) from Pseudomonas aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bacterioferritin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
8QYF
 
 | |
6U9M
 
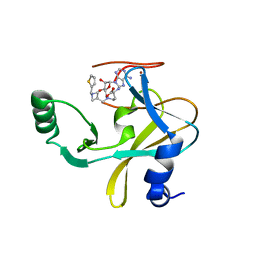 | | MLL1 SET N3861I/Q3867L bound to inhibitor 16 (TC-5109) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(thiophen-2-yl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
8CJZ
 
 | | Carin1 bacteriophage mature capsid | | Descriptor: | Capsid Decoration Protein, Major Capsid Protein, Spike Base Protein | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
9AYL
 
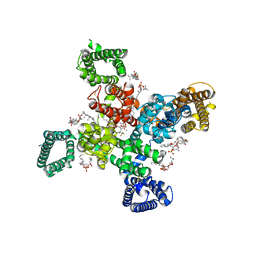 | | Cryo-EM structure of human Cav3.2 with ACT-709478 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-08 | | Release date: | 2024-04-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
7RBR
 
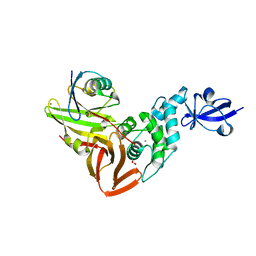 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Papain-like protease, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
6XWB
 
 | |
