6VB5
 
 | |
6SIY
 
 | | PaaK family AMP-ligase with AMP and substrate | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYANTHRANILIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7O2L
 
 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
8SPW
 
 | | PS3 F1 Rotorless, low ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
7LMP
 
 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 36 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-8-(1H-imidazole-4-carbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione] | | Descriptor: | 3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-8-(1~{H}-imidazol-5-ylcarbonyl)-1,3,8-triazaspiro[4.5]decane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7LMO
 
 | | Structure of full-length human lambda-6A light chain JTO in complex with stabilizer 34 [3-(2-(7-(diethylamino)-4-methyl-2-oxo-2H-chromen-3-yl)ethyl)-7-(1H-imidazole-5-carbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione] | | Descriptor: | (5~{R})-3-[2-[7-(diethylamino)-4-methyl-2-oxidanylidene-chromen-3-yl]ethyl]-7-(1~{H}-imidazol-4-ylcarbonyl)-1,3,7-triazaspiro[4.4]nonane-2,4-dione, JTO light chain, PHOSPHATE ION | | Authors: | Yan, N.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Coumarin-Based Kinetic Stabilizers of Amyloidogenic Immunoglobulin Light Chains Using Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
6X78
 
 | |
6UZP
 
 | |
6SXI
 
 | | Antibody-anti-idiotype complex: AP33 Fab (hepatitis C virus E2 antibody) - B2.1A scFv (anti-idiotype) | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Taylor, G.L, Potter, J.A, Fadda, V, Patel, A.H, Owsianka, A.M, Cowtan, V.M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of a structural epitope mimic: an idiotypic approach to HCV vaccine design.
NPJ Vaccines, 6, 2021
|
|
7V8W
 
 | | Crystal structure of PsEst3 S128A variant complexed with malonate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MALONIC ACID, ... | | Authors: | Son, J, Kim, H, Kim, H.W. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical insights into PsEst3, a new GHSR-type esterase obtained from Paenibacillus sp. R4.
Iucrj, 10, 2023
|
|
6D9F
 
 | | Protein 60 with aldehyde deformylating oxidase activity from Kitasatospora setae | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Putative VlmB homolog, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2018-04-28 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Biochemistry, 59, 2020
|
|
6XAM
 
 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-homoalanine) | | Descriptor: | (3S,6S)-3-ethyl-6-[(1H-indol-3-yl)methyl]piperazine-2,5-dione, 1,2-ETHANEDIOL, NzeB, ... | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
8H5V
 
 | | Crystal structure of the FleQ domain of Vibrio cholerae FlrA | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, flagellar regulatory protein A | | Authors: | Dasgupta, J, Chakraborty, S. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The N-terminal FleQ domain of the Vibrio cholerae flagellar master regulator FlrA plays pivotal structural roles in stabilizing its active state.
Febs Lett., 597, 2023
|
|
6XCT
 
 | | Porcine pepsin in complex with amprenavir | | Descriptor: | Pepsin A, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Porcine pepsin in complex with amprenavir
To Be Published
|
|
7Q9Q
 
 | |
7OKT
 
 | | X-ray structure of soluble EPCR in C2221 space group | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Endothelial protein C receptor, ... | | Authors: | Erausquin, E, Dichiara, M.G, Lopez-Sagaseta, J. | | Deposit date: | 2021-05-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of a broad lipid repertoire associated to the endothelial cell protein C receptor (EPCR).
Sci Rep, 12, 2022
|
|
6XD2
 
 | | Porcine pepsin in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Pepsin A | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Porcine pepsin in complex with darunavir
To Be Published
|
|
8GZ6
 
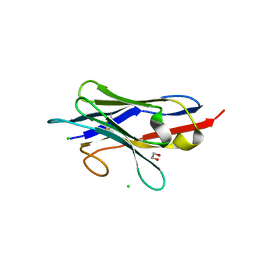 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nanobody P17 | | Authors: | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | Deposit date: | 2022-09-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
4QHS
 
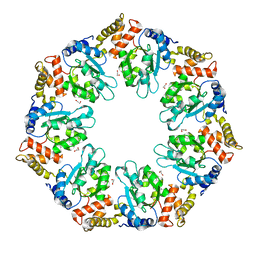 | | Crystal structure of AAA+sigma 54 activator domain of the flagellar regulatory protein FlrC of Vibrio cholerae in nucleotide free state | | Descriptor: | 1,2-ETHANEDIOL, Flagellar regulatory protein C | | Authors: | Dey, S, Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC
J.Biol.Chem., 290, 2015
|
|
8SPK
 
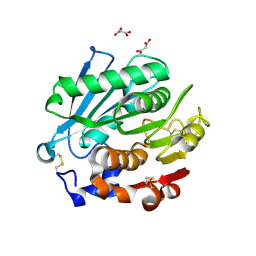 | | Crystal structure of Antarctic PET-degrading enzyme | | Descriptor: | Lipase 1, MALONATE ION | | Authors: | Furtado, A.A, Blazquez-Sanchez, P, Grinen, A, Vargas, J.A, Leonardo, D.A, Sculaccio, S.A, Pereira, H.M, Diez, B, Garratt, R.C, Ramirez-Sarmiento, C.A. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the catalytic activity of an Antarctic PET-degrading enzyme by loop exchange.
Protein Sci., 32, 2023
|
|
5MYE
 
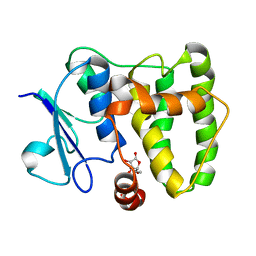 | |
9NSR
 
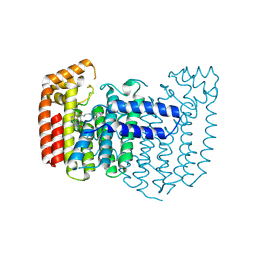 | |
7T91
 
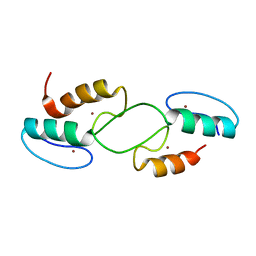 | | Crystal structure of Zinc finger motif 1 and 2 of GLI1 DNA binding region | | Descriptor: | Isoform 2 of Zinc finger protein GLI1, ZINC ION | | Authors: | Wu, M, Zhang, S, Augelli-Szanfran, C.E, Boohaker, R.J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure Characterization of Zinc Finger Motif 1 and 2 of GLI1 DNA Binding Region
Int J Mol Sci, 25, 2024
|
|
1KBD
 
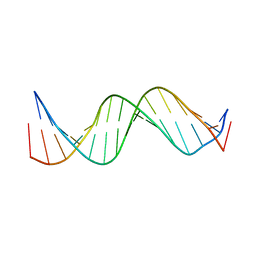 | | SOLUTION STRUCTURE OF A 16 BASE-PAIR DNA RELATED TO THE HIV-1 KAPPA B SITE | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*AP*GP*G)-3') | | Authors: | Tisne, C, Hantz, E, Hartmann, B, Delepierre, M. | | Deposit date: | 1998-11-28 | | Release date: | 1998-12-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a non-palindromic 16 base-pair DNA related to the HIV-1 kappa B site: evidence for BI-BII equilibrium inducing a global dynamic curvature of the duplex.
J.Mol.Biol., 279, 1998
|
|
7Z8E
 
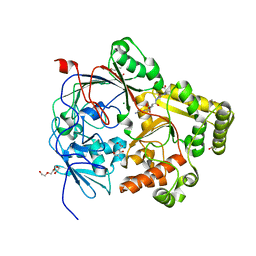 | | Crystal structure of the substrate-binding protein YejA from S. meliloti in complex with peptide fragment | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, GLY-SER-ASP-VAL-ALA, ... | | Authors: | Morera, S, Vigouroux, V, Travin, D.Y. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual-Uptake Mode of the Antibiotic Phazolicin Prevents Resistance Acquisition by Gram-Negative Bacteria.
Mbio, 14, 2023
|
|
