3K7S
 
 | |
8DSB
 
 | | Lambda Bacteriophage Orf63 | | Descriptor: | Xis (Excision72) | | Authors: | Donaldson, L.W. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Bad Phages in Good Bacteria: Role of the Mysterious orf63 of lambda and Shiga Toxin-Converting Phi 24 B Bacteriophages.
Front Microbiol, 8, 2017
|
|
158D
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF C-C-A-A-G-C-T-T-G-G AND ITS IMPLICATIONS FOR BENDING IN B-DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*GP*CP*TP*TP*GP*G)-3') | | Authors: | Grzeskowiak, K, Goodsell, D.S, Kaczor-Grzeskowiak, M, Cascio, D, Dickerson, R.E. | | Deposit date: | 1994-02-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of C-C-A-A-G-C-T-T-G-G and its implications for bending in B-DNA.
Biochemistry, 32, 1993
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
7UNT
 
 | |
7YG3
 
 | | Crystal structure of HLA-B*13:01 | | Descriptor: | ARG-GLN-ASP-ILE-LEU-ASP-LEU-TRP-ILE, Beta-2-microglobulin, MHC class I antigen | | Authors: | Wang, H.S, Ouyang, S.Y. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional and structural characteristics of HLA-B*13:01-mediated specific T cells reaction in dapsone-induced drug hypersensitivity.
J.Biomed.Sci., 29, 2022
|
|
7UD0
 
 | | Room Temperature Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Schneps, C.M, Crane, B.R. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Room-temperature serial synchrotron crystallography of Drosophila cryptochrome.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6TU9
 
 | | The ROR1 Pseudokinase Domain Bound To Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Inactive tyrosine-protein kinase transmembrane receptor ROR1 | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Niininen, W, Ungureanu, D, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
1HE4
 
 | | Human biliverdin IX beta reductase: NADP/FMN ternary complex | | Descriptor: | BILIVERDIN IX BETA REDUCTASE, FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pereira, P.J.B, Macedo-Ribeiro, S, Parraga, A, Perez-Luque, R, Cunningham, O, Darcy, K, Mantle, T.J, Coll, M. | | Deposit date: | 2000-11-19 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Human Biliverdin Ix Beta Reductase, an Early Fetal Bilirubin Ix Producing Enzyme
Nat.Struct.Biol., 8, 2001
|
|
1GPA
 
 | |
4QYH
 
 | | CHK1 kinase domain in complex with diazacarbazole GNE-783 | | Descriptor: | 3-[4-(4-methylpiperazin-1-yl)phenyl]-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Wiesmann, C, Wu, P. | | Deposit date: | 2014-07-24 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of the 1,7-diazacarbazole class of inhibitors of checkpoint kinase 1.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1HE3
 
 | | Human biliverdin IX beta reductase: NADP/mesobiliverdin IV alpha ternary complex | | Descriptor: | BILIVERDIN IX BETA REDUCTASE, MESOBILIVERDIN IV ALPHA, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pereira, P.J.B, Macedo-Ribeiro, S, Parraga, A, Perez-Luque, R, Cunningham, O, Darcy, K, Mantle, T.J, Coll, M. | | Deposit date: | 2000-11-18 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Human Biliverdin Ix Beta Reductase, an Early Fetal Bilirubin Ix Producing Enzyme
Nat.Struct.Biol., 8, 2001
|
|
1GXS
 
 | | Crystal Structure of Hydroxynitrile Lyase from Sorghum bicolor in Complex with Inhibitor Benzoic Acid: a novel cyanogenic enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZOIC ACID, DECANOIC ACID, ... | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Wajant, H, Effenberger, F. | | Deposit date: | 2002-04-11 | | Release date: | 2002-10-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Hydroxynitrile Lyase from Sorghum Bicolor in Complex with the Inhibitor Benzoic Acid: A Novel Cyanogenic Enzyme
Biochemistry, 41, 2002
|
|
3JUM
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with 5-bromo-2-((1S,3R)-3-carboxycyclohexylamino)benzoic acid | | Descriptor: | 5-bromo-2-{[(1S,3R)-3-carboxycyclohexyl]amino}benzoic acid, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2INS
 
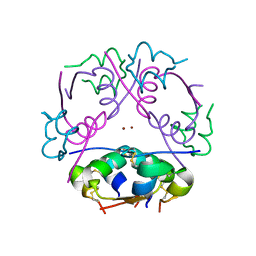 | | THE STRUCTURE OF DES-PHE B1 BOVINE INSULIN | | Descriptor: | DES-PHE B1 INSULIN (CHAIN A), DES-PHE B1 INSULIN (CHAIN B), ZINC ION | | Authors: | Smith, G.D, Duax, W.L, Dodson, E.J, Dodson, G.G, Degraaf, R.A.G, Reynolds, C.D. | | Deposit date: | 1982-05-10 | | Release date: | 1982-08-05 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of Des-Phe B1 Bovine Insulin
Acta Crystallogr.,Sect.B, 38, 1982
|
|
194D
 
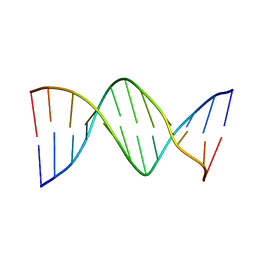 | | X-RAY STRUCTURES OF THE B-DNA DODECAMER D(CGCGTTAACGCG) WITH AN INVERTED CENTRAL TETRANUCLEOTIDE AND ITS NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*AP*AP*CP*GP*CP*G)-3') | | Authors: | Balendiran, K, Rao, S.T, Sekharudu, C.Y, Zon, G, Sundaralingam, M. | | Deposit date: | 1994-10-04 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the B-DNA dodecamer d(CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1VGC
 
 | | GAMMA-CHYMOTRYPSIN L-PARA-CHLORO-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | GAMMA CHYMOTRYPSIN, L-1-(4-CHLOROPHENYL)-2-(ACETAMIDO)ETHANE BORONIC ACID, SULFATE ION | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
7ML0
 
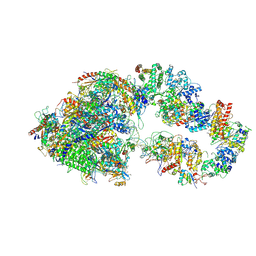 | | RNA polymerase II pre-initiation complex (PIC1) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML1
 
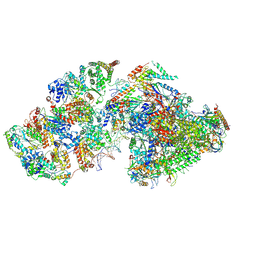 | | RNA polymerase II pre-initiation complex (PIC2) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML2
 
 | | RNA polymerase II pre-initiation complex (PIC3) | | Descriptor: | BJ4_G0004860.mRNA.1.CDS.1, BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
4AIL
 
 | | Crystal Structure of an Evolved Replicating DNA Polymerase | | Descriptor: | 5'-D(*AP*CP*GP*GP*GP*TP*AP*AP*GP*CP*AP)-3', 5'-D(*TP*GP*CP*TP*TP*AP*CP*DOCP)-3', DNA POLYMERASE | | Authors: | Wynne, S.A, Holliger, P, Leslie, A.G.W. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of an Apo and a Binary Complex of an Evolved Archeal B Family DNA Polymerase Capable of Synthesising Highly Cy-Dye Labelled DNA.
Plos One, 8, 2013
|
|
1YRJ
 
 | | Crystal Structure of Apramycin bound to a Ribosomal RNA A site oligonucleotide | | Descriptor: | APRAMYCIN, Bacterial 16 S Ribosomal RNA A Site Oligonucleotide, MAGNESIUM ION, ... | | Authors: | Han, Q, Zhao, Q, Fish, S, Simonsen, K.B, Vourloumis, D, Froelich, J.M, Wall, D, Hermann, T. | | Deposit date: | 2005-02-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition by glycoside pseudo base pairs and triples in an apramycin-RNA complex.
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
8EGQ
 
 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2S)-2-ethyl-4-{[(2'M)-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazine, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
1HDO
 
 | | Human biliverdin IX beta reductase: NADP complex | | Descriptor: | BILIVERDIN IX BETA REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pereira, P.J.B, Macedo-Ribeiro, S, Parraga, A, Perez-Luque, R, Cunningham, O, Darcy, K, Mantle, T.J, Coll, M. | | Deposit date: | 2000-11-16 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of Human Biliverdin Ix Beta Reductase, an Early Fetal Bilirubin Ix Producing Enzyme
Nat.Struct.Biol., 8, 2001
|
|
