6X63
 
 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7R3Z
 
 | | Tankyrase 2 catalytic domain in complex with OUL40 | | Descriptor: | 6-methyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, GLYCEROL, Poly [ADP-ribose] polymerase tankyrase-2, ... | | Authors: | Murthy, S, Venkannagari, H, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
9C7I
 
 | | Crystal structure of caryolan-1-ol synthase from S. griseus with PEG molecule in the active site | | Descriptor: | (+)-caryolan-1-ol synthase, CALCIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2024-06-10 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of Caryolan-1-ol Synthase, a Sesquiterpene Synthase Catalyzing an Initial Anti-Markovnikov Cyclization Reaction.
Biochemistry, 63, 2024
|
|
7R3T
 
 | | Crystal structure of the Dimeric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Beta-1,3-glucanase bglH, CHLORIDE ION, ... | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Paenibacillus illinoisensis IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
6Z4Q
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule inverse agonist SR142948A | | Descriptor: | 2-[[5-(2,6-dimethoxyphenyl)-1-[4-[3-(dimethylamino)propyl-methyl-carbamoyl]-2-propan-2-yl-phenyl]pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6ZIN
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-26 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z4S
 
 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
9OOI
 
 | | Crystal structure of dihydrofolate reductase (DHFR) from the filarial nematode W. bancrofti in complex with NADPH and antifolate 2-({4-[(2-amino-4-oxo-4,7-dihydro-1H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)benzoic acid (OG7 or TSD001) | | Descriptor: | 2-({4-[(2-amino-4-oxo-4,7-dihydro-1H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)benzoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dihydrofolate reductase | | Authors: | Frey, K.M, Goodey, N.M, Kwarteng, S. | | Deposit date: | 2025-05-15 | | Release date: | 2025-08-27 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A virtual screening strategy to repurpose antifolate compounds as W.bancrofti DHFR inhibitors.
Bioorg.Med.Chem.Lett., 129, 2025
|
|
6XFV
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C IN COMPLEX WITH A NOVEL INVERSE AGONIST | | Descriptor: | 1-(4-{(3S,4S)-4-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-3-methyl-3-phenylpyrrolidine-1-carbonyl}piperidin-1-yl)ethan-1-one, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-06-16 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of (3S,4S)-3-methyl-3-(4-fluorophenyl)-4-(4-(1,1,1,3,3,3-hexafluoro-2-hydroxyprop-2-yl)phenyl)pyrrolidines as novel ROR gamma t inverse agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6ZF8
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[2-hydroxy-2-oxoethyl-(3-methoxyphenyl)sulfonyl-amino]phenyl]-5-[(1~{S},2~{S})-2-phenylcyclopropyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF5
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[(4-butylphenyl)sulfonyl-(2-hydroxy-2-oxoethyl)amino]phenyl]-5-cyclopropyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6PNN
 
 | |
8RAQ
 
 | | Crystal structure of Mycobacterium tuberculosis MmaA1 with S-adenosyl methionine (SAM) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mycolic acid methyltransferase MmaA1, ... | | Authors: | Chaudhary, B, Kobakhidze, G, Wachelder, L, Mazumdar, P.A, Madhurantakam, C, Dong, G. | | Deposit date: | 2023-12-01 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the mycolic acid methyl transferase 1 (MmaA1) from Mycobacterium tuberculosis in the apo-form and in complex with different cofactors reveal unique features for substrate binding.
J.Biomol.Struct.Dyn., 2025
|
|
8F1C
 
 | | Voltage-gated potassium channel Kv3.1 with novel positive modulator (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one (compound 4) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one, POTASSIUM ION, ... | | Authors: | Chen, Y, Ishchenko, A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Identification and structural and biophysical characterization of a positive modulator of human Kv3.1 channels.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8R0J
 
 | | Crystal structure of the retromer complex VPS29/VPS35 with the ligand bis-1,3-phenyl guanylhydrazone, 2a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bis-1,3-phenyl guanylhydrazon, Vacuolar protein sorting-associated protein 29, ... | | Authors: | Milani, M, Fagnani, E. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Stabilization of the retromer complex: Analysis of novel binding sites of bis-1,3-phenyl guanylhydrazone 2a to the VPS29/VPS35 interface.
Comput Struct Biotechnol J, 23, 2024
|
|
8EKX
 
 | | Structure of MBP-Mcl-1 in complex with MIK665 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy]phenyl]propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
7MEE
 
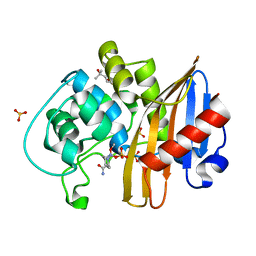 | | CDD-1 beta-lactamase in imidazole/MPD 6 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEC
 
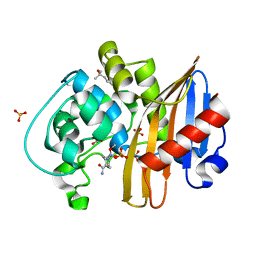 | | CDD-1 beta-lactamase in imidazole/MPD 4 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEF
 
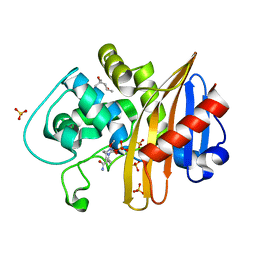 | | CDD-1 beta-lactamase in imidazole/MPD 10 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MED
 
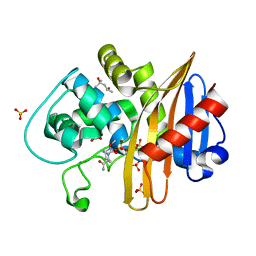 | | CDD-1 beta-lactamase in imidazole/MPD 5 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEB
 
 | | CDD-1 beta-lactamase in imidazole/MPD 2 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
6ZIC
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH 3S-HYDROXYBUTANOYL-COA AND NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-HYDROXYBUTANOYL-COENZYME A, GLYCEROL, ... | | Authors: | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | Deposit date: | 2020-06-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural enzymology studies with the substrate 3S-hydroxybutanoyl-CoA: bifunctional MFE1 is a less efficient dehydrogenase than monofunctional HAD.
Febs Open Bio, 14, 2024
|
|
6VKO
 
 | |
6Z2V
 
 | | CLK3 A319V mutant bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, ... | | Authors: | Schroeder, M, Chaikuad, A, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6PNM
 
 | |
