7XX5
 
 | | Crystal Structure of Nucleosome-H1.3 Linker Histone Assembly (sticky-169a DNA fragment) | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H1.3, ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169a DNA fragment)
To Be Published
|
|
6AY2
 
 | | Structure of CathB with covalently linked Compound 28 | | Descriptor: | Cathepsin B, N~1~-[(2S)-1-amino-5-(carbamoylamino)pentan-2-yl]-N'~1~-[(1R)-1-(thiophen-3-yl)ethyl]cyclobutane-1,1-dicarboxamide | | Authors: | Kiefer, J.R, Steinbacher, S. | | Deposit date: | 2017-09-07 | | Release date: | 2017-12-27 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Peptidomimetic Antibody-Drug Conjugate Linkers with Enhanced Protease Specificity.
J. Med. Chem., 61, 2018
|
|
1L55
 
 | |
5SZP
 
 | | Protocadherin Gamma B7 extracellular cadherin domains 1-4 P21 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin Gamma B7, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
4QRR
 
 | | Crystal Structure of HLA B*3501-IPS in complex with a Delta-Beta TCR, clone 12 TCR | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Chabrol, E, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
1UZU
 
 | | Glycogen Phosphorylase b in complex with indirubin-5'-sulphonate | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Kosmopoulou, M.N, Leonidas, D.D, Chrysina, E.D, Bischler, N, Eisenbrand, G, Sakarellos, C.E, Pauptit, R, Oikonomakos, N.G. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of the potential antitumour agent indirubin-5-sulphonate at the inhibitor site of rabbit muscle glycogen phosphorylase b. Comparison with ligand binding to pCDK2-cyclin A complex.
Eur. J. Biochem., 271, 2004
|
|
7Y6K
 
 | |
1L61
 
 | |
1O6I
 
 | | Chitinase B from Serratia marcescens complexed with the catalytic intermediate mimic cyclic dipeptide CI4. | | Descriptor: | Chitinase, GLYCEROL, SULFATE ION, ... | | Authors: | Houston, D.R, Eggleston, I, Synstad, B, Eijsink, V.G.H, van Aalten, D.M.F. | | Deposit date: | 2002-10-03 | | Release date: | 2003-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The cyclic dipeptide CI-4 [cyclo-(l-Arg-d-Pro)] inhibits family 18 chitinases by structural mimicry of a reaction intermediate.
Biochem. J., 368, 2002
|
|
5CSJ
 
 | | S100B-RSK1 crystal structure B | | Descriptor: | CALCIUM ION, CHLORIDE ION, Protein S100-B, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Ribosomal S6 Kinase 1 (RSK1) Inhibition by S100B Protein: MODULATION OF THE EXTRACELLULAR SIGNAL-REGULATED KINASE (ERK) SIGNALING CASCADE IN A CALCIUM-DEPENDENT WAY.
J.Biol.Chem., 291, 2016
|
|
6R7B
 
 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 1 | | Descriptor: | CRISPR-associated (Cas) DxTHG family, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Molina, R, Montoya, G, Sofos, N, Stella, S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
6F7T
 
 | |
5UYT
 
 | | Crystal structure of ice binding protein from an Antarctic bacterium Flavobacteriaceae | | Descriptor: | Ice-binding protein, NITRATE ION | | Authors: | Wang, C, Pakhomova, S, Newcomer, M.E, Christner, B.C, Luo, B.-H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of antifreeze activity of a bacterial multi-domain antifreeze protein.
PLoS ONE, 12, 2017
|
|
5IYQ
 
 | |
1GQH
 
 | | Quercetin 2,3-dioxygenase in complex with the inhibitor kojic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXY-2-(HYDROXYMETHYL)-4H-PYRAN-4-ONE, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional Analysis of the Copper-Dependent Quercetin 2,3-Dioxygenase.1.Ligand-Induced Coordination Changes Probed by X-Ray Crystallography: Inhibition, Ordering Effect and Mechanistic Insights
Biochemistry, 41, 2002
|
|
2XH6
 
 | | Clostridium perfringens enterotoxin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, HEAT-LABILE ENTEROTOXIN B CHAIN, octyl beta-D-glucopyranoside | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, MCClane, B.A, Basak, A.K. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
6EWC
 
 | | Crystal structure of non-phosphorylated form of RLS PHOSPHOPEPTIDE BOUND TO HLA-A2 in complex with LILRB1 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Mohammed, F, Stones, D.H, Willcox, B.E. | | Deposit date: | 2017-11-03 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Application of the immunoregulatory receptor LILRB1 as a crystallisation chaperone for human class I MHC complexes.
J. Immunol. Methods, 464, 2019
|
|
6SHB
 
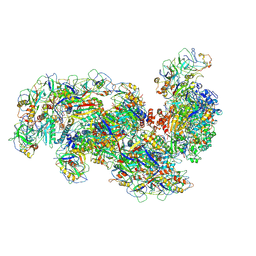 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1, in the presence of ssDNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
5GN9
 
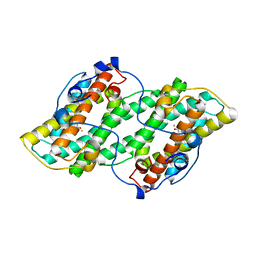 | | Crystal structure of alternative oxidase from Trypanosoma brucei brucei complexed with cumarin derivative-17b | | Descriptor: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2016-07-19 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
8PJ3
 
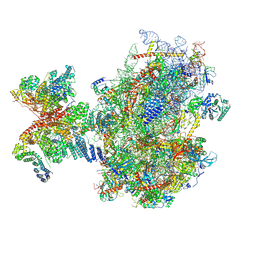 | | Structure of human 48S translation initiation complex upon transfer of initiator tRNA to eIF5B (48S-3) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
8PJ1
 
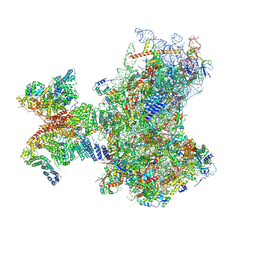 | | Structure of human 48S translation initiation complex in open codon scanning state (48S-1) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
8PJ6
 
 | | Structure of human 48S translation initiation complex with initiator tRNA, eIF1A and eIF3 (off-pathway) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
8PJ5
 
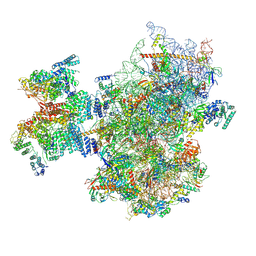 | | Structure of human 48S translation initiation complex after eIF2 release prior 60S subunit joining (48S-5) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
8PJ4
 
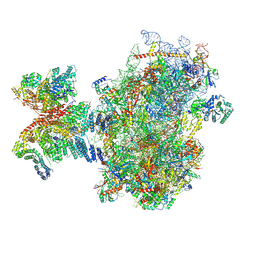 | | Structure of human 48S translation initiation complex after eIF5 release (48S-4) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
5CX8
 
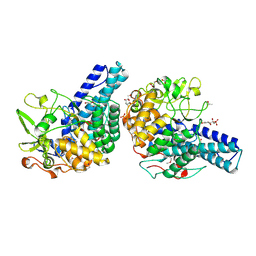 | | Structure of RagB, a major immunodominant virulence factor of Porphyromonas gingivalis. | | Descriptor: | 3-deoxy-5-O-phosphono-beta-D-ribofuranose, 3-deoxy-beta-D-glucopyranose, 6-O-phosphono-D-tagatose, ... | | Authors: | Goulas, T, Garcia-Ferrer, I, Hutcherson, J.A, Potempa, B.A, Potempa, J, Scott, D.A, Gomis-Ruth, F.X. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of RagB, a major immunodominant outer-membrane surface receptor antigen of Porphyromonas gingivalis.
Mol Oral Microbiol, 31, 2016
|
|
