6EI5
 
 | |
4EV6
 
 | | The complete structure of CorA magnesium transporter from Methanocaldococcus jannaschii | | Descriptor: | MAGNESIUM ION, Magnesium transport protein CorA, UNDECYL-MALTOSIDE | | Authors: | Guskov, A, Nordin, N, Reynaud, A, Engman, H, Lundback, A.-K, Jong, A.J.O, Cornvik, T, Phua, T, Eshaghi, S. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the mechanisms of Mg2+ uptake, transport, and gating by CorA
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6EN2
 
 | |
7NP4
 
 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NP3
 
 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
6E6N
 
 | | Pheromone from Euplotes raikovi, Er-13 | | Descriptor: | Pheromone from Euplotes raikovi Er-13 | | Authors: | Finke, A.D, Marsh, M.E. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-07 | | Method: | X-RAY DIFFRACTION (1.363 Å) | | Cite: | Ab initio crystal structure determination of Euplotes raikovi pheromones from high-resolution data
To Be Published
|
|
6E7G
 
 | |
8PQG
 
 | |
8Q5B
 
 | |
8Q79
 
 | | Structure of mBaoJin at pH 6.5 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Nikolaeva, A.Y, Borshchevsky, V, Qin, W, Subach, F.V. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
6EAL
 
 | |
6EIZ
 
 | | A de novo designed hexameric coiled coil CC-Hex2 with farnesol bound in the channel. | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, CC-Hex2 | | Authors: | Rhys, G.G, Burton, A.J, Dawson, W.M, Thomas, F, Woolfson, D.N. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De Novo-Designed alpha-Helical Barrels as Receptors for Small Molecules.
ACS Synth Biol, 7, 2018
|
|
6EN3
 
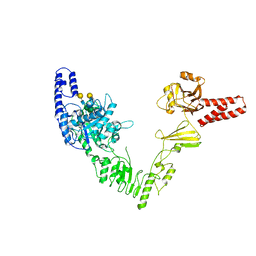 | | Crystal structure of full length EndoS from Streptococcus pyogenes in complex with G2 oligosaccharide. | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2,Multifunctional-autoprocessing repeats-in-toxin, NICKEL (II) ION, ... | | Authors: | Trastoy, B, Klontz, E.H, Orwenyo, J, Marina, A, Wang, L.X, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2017-10-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural basis for the recognition of complex-type N-glycans by Endoglycosidase S.
Nat Commun, 9, 2018
|
|
6ENQ
 
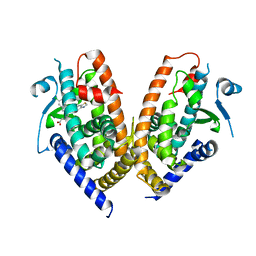 | | Structure of human PPAR gamma LBD in complex with Lanifibranor (IVA337) | | Descriptor: | 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Boubia, B, Poupardin, O, Barth, M, Amaudrut, J, Broqua, P, Tallandier, M, Zeyer, D. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, Synthesis, and Evaluation of a Novel Series of Indole Sulfonamide Peroxisome Proliferator Activated Receptor (PPAR) alpha / gamma / delta Triple Activators: Discovery of Lanifibranor, a New Antifibrotic Clinical Candidate.
J. Med. Chem., 61, 2018
|
|
6ENZ
 
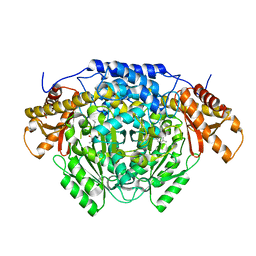 | | Crystal structure of mouse GADL1 | | Descriptor: | Acidic amino acid decarboxylase GADL1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Raasakka, A, Mahootchi, E, Winge, I, Luan, W, Kursula, P, Haavik, J. | | Deposit date: | 2017-10-07 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the mouse acidic amino acid decarboxylase GADL1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7N6W
 
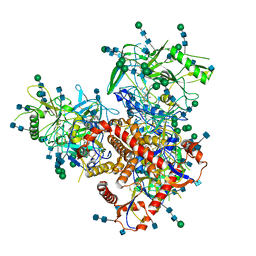 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U2 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
4OS0
 
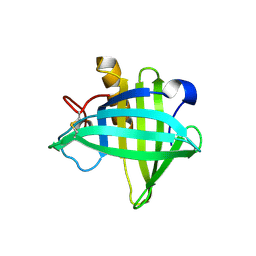 | | Three-dimensional structure of the C65A-W54F double mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7NMH
 
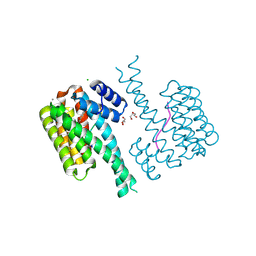 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-070 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) methanesulfonate, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
6EOO
 
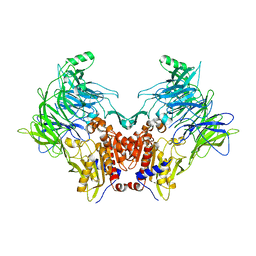 | | DPP8 - Apo, space group 20 | | Descriptor: | Dipeptidyl peptidase 8 | | Authors: | Ross, B.R, Huber, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7N6U
 
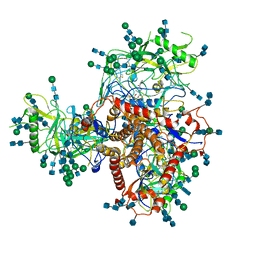 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U1 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
6EN1
 
 | |
8Q5Q
 
 | | d(ATTTC)3 dimeric structure | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*(DNR)P*AP*TP*TP*TP*CP*AP*TP*TP*TP*C)-3') | | Authors: | Trajkovski, M, Pastore, A, Plavec, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Dimeric structures of DNA ATTTC repeats promoted by divalent cations.
Nucleic Acids Res., 52, 2024
|
|
6ES7
 
 | |
6E6O
 
 | | Pheromone from Euplotes raikovi, Er-1 | | Descriptor: | Mating pheromone Er-1/Er-3 | | Authors: | Finke, A.D, Marsh, M.E. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-07 | | Method: | X-RAY DIFFRACTION (0.7 Å) | | Cite: | Ab initio crystal structure determination of Euplotes raikovi pheromones from high-resolution data
To Be Published
|
|
6EAI
 
 | |
