6ND4
 
 | | Conformational switches control early maturation of the eukaryotic small ribosomal subunit | | Descriptor: | 18S rRNA 5' domain start, 5'ETS rRNA, Bud21, ... | | Authors: | Hunziker, M, Barandun, J, Klinge, S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Conformational switches control early maturation of the eukaryotic small ribosomal subunit.
Elife, 8, 2019
|
|
5MMD
 
 | | TMB-1. Structural insights into TMB-1 and the role of residue 119 and 228 in substrate and inhibitor binding | | Descriptor: | CHLORIDE ION, Metallo-beta-lactamase 1, ZINC ION | | Authors: | Skagseth, S, Christopeit, T, Akhter, S, Bayer, A, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2016-12-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into TMB-1 and the Role of Residues 119 and 228 in Substrate and Inhibitor Binding.
Antimicrob. Agents Chemother., 61, 2017
|
|
4IRL
 
 | | X-ray structure of the CARD domain of zebrafish GBP-NLRP1 like protein | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jin, T, Huang, M, Smith, P, Xiao, T. | | Deposit date: | 2013-01-15 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of the caspase-recruitment domain from a zebrafish guanylate-binding protein.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3F5F
 
 | | Crystal structure of heparan sulfate 2-O-sulfotransferase from gallus gallus as a maltose binding protein fusion. | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1, ... | | Authors: | Bethea, H.N, Xu, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2008-11-03 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
8JYX
 
 | |
1CDR
 
 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 | | Authors: | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | Deposit date: | 1994-06-01 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
6O6D
 
 | |
1ANF
 
 | | MALTODEXTRIN BINDING PROTEIN WITH BOUND MALTOSE | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Spurlino, J.C, Quiocho, F.A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor.
Structure, 5, 1997
|
|
7MN6
 
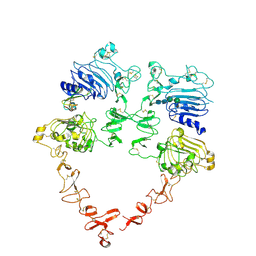 | | Structure of the HER2 S310F/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7MN5
 
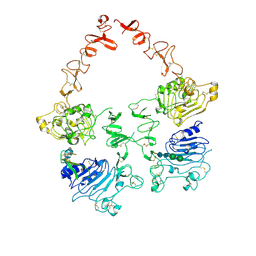 | | Structure of the HER2/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7MN8
 
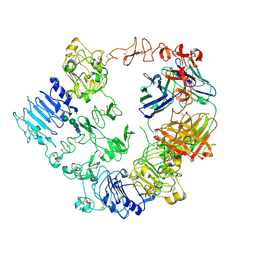 | | Structure of the HER2/HER3/NRG1b Heterodimer Extracellular Domain bound to Trastuzumab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
7M74
 
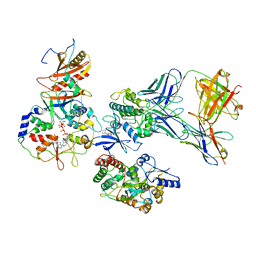 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
1CDQ
 
 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | Descriptor: | CD59 | | Authors: | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | Deposit date: | 1994-06-01 | | Release date: | 1994-09-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
4JBZ
 
 | | Structure of Mcm10 coiled-coil region | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN FUSED WITH XENOPUS LAEVIS MCM10 COILED-COIL REGION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Du, W, Adhikary, S, Eichman, B.F. | | Deposit date: | 2013-02-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mcm10 self-association is mediated by an N-terminal coiled-coil domain.
Plos One, 8, 2013
|
|
4H1G
 
 | | Structure of Candida albicans Kar3 motor domain fused to maltose-binding protein | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Allingham, J.A, Duan, D, Delorme, C, Joshi, M. | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the Candida albicans Kar3 kinesin motor domain fused to maltose-binding protein.
Biochem.Biophys.Res.Commun., 428, 2012
|
|
3G7W
 
 | | Islet Amyloid Polypeptide (IAPP or Amylin) Residues 1 to 22 fused to Maltose Binding Protein | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | Authors: | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
7MHW
 
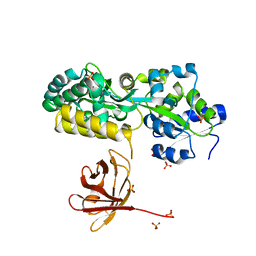 | | Crystal structure of the protease inhibitor U-Omp19 from Brucella abortus fused to Maltose-binding protein | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Outer membrane lipoprotein omp19, SULFATE ION | | Authors: | Darriba, M.L, Klinke, S, Otero, L.H, Cerutti, M.L, Cassataro, J, Pasquevich, K.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A disordered region retains the full protease inhibitor activity and the capacity to induce CD8 + T cells in vivo of the oral vaccine adjuvant U-Omp19.
Comput Struct Biotechnol J, 20, 2022
|
|
3G7V
 
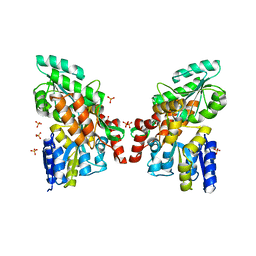 | | Islet Amyloid Polypeptide (IAPP or Amylin) fused to Maltose Binding Protein | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | Authors: | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2009-02-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
4JKM
 
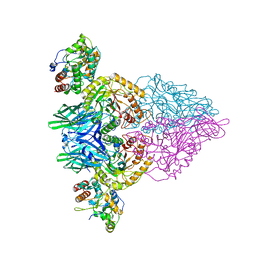 | |
6OB5
 
 | | Computationally-designed, modular sense/response system (S3-2D) | | Descriptor: | Ankyrin Repeat Domain (AR), S3-2D variant, FARNESYL DIPHOSPHATE, ... | | Authors: | Thompson, M.C, Glasgow, A.A, Huang, Y.M, Fraser, J.S, Kortemme, T. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Computational design of a modular protein sense-response system.
Science, 366, 2019
|
|
7MQ7
 
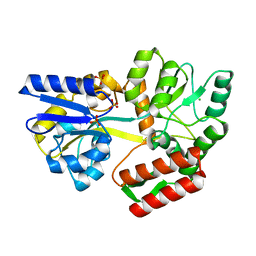 | | Tetragonal Maltose Binding Protein | | Descriptor: | CHLORIDE ION, Maltodextrin-binding protein, SULFATE ION, ... | | Authors: | Thaker, A, Sirajudeen, L, Simmons, C.R, Nannenga, B.L. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided identification of a peptide for bio-enabled gold nanoparticle synthesis.
Biotechnol.Bioeng., 118, 2021
|
|
7MQ6
 
 | | Tetragonal Maltose Binding Protein in the presence of gold | | Descriptor: | CHLORIDE ION, GOLD ION, Maltodextrin-binding protein, ... | | Authors: | Thaker, A, Sirajudeen, L, Simmons, C.R, Nannenga, B.L. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | Structure-guided identification of a peptide for bio-enabled gold nanoparticle synthesis.
Biotechnol.Bioeng., 118, 2021
|
|
5LEL
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_10_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | Descriptor: | DD_Off7_10_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LOF
 
 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent inhibitor of MCL1 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-[2,2,2-tris(fluoranyl)ethyl]pyrazol-3-yl]methoxy]phenyl]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Kotschy, A, Szlavik, Z, Murray, J, Davidson, J, Csekei, M, Paczal, A, Szabo, Z, Sipos, S, Radics, G, Proszenyak, A, Balint, B, Ondi, L, Blasko, G, Robertson, A, Surgenor, A, Chen, I, Matassova, N, Smith, J, Pedder, C, Graham, C, Geneste, O. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models.
Nature, 538, 2016
|
|
