3K3K
 
 | | Crystal structure of dimeric abscisic acid (ABA) receptor pyrabactin resistance 1 (PYR1) with ABA-bound closed-lid and ABA-free open-lid subunits | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism of abscisic acid binding and signaling by dimeric PYR1.
Science, 326, 2009
|
|
1OVK
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with N-Allyl-Aniline | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-26 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
3K01
 
 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | Descriptor: | Acarbose/maltose binding protein GacH, SULFATE ION | | Authors: | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | Deposit date: | 2009-09-24 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
2NUZ
 
 | | crystal structure of alpha spectrin SH3 domain measured at room temperature | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Agarwal, V, Faelber, K, Hologne, M, Chevelkov, V, Oschkinat, H, Diehl, A, Reif, B. | | Deposit date: | 2006-11-10 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of MAS solid-state NMR and X-ray crystallographic data in the analysis of protein dynamics in the solid state
To be Published
|
|
3K4T
 
 | |
1QK1
 
 | | CRYSTAL STRUCTURE OF HUMAN UBIQUITOUS MITOCHONDRIAL CREATINE KINASE | | Descriptor: | CREATINE KINASE, UBIQUITOUS MITOCHONDRIAL, PHOSPHATE ION | | Authors: | Eder, M, Schlattner, U, Fritz-Wolf, K, Wallimann, T, Kabsch, W. | | Deposit date: | 1999-07-08 | | Release date: | 2000-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Ubiquitous Mitochondrial Creatine Kinase
Proteins: Struct.,Funct., Genet., 39, 2000
|
|
3B4R
 
 | | Site-2 Protease from Methanocaldococcus jannaschii | | Descriptor: | Putative zinc metalloprotease MJ0392, ZINC ION | | Authors: | Jeffrey, P.D, Feng, L, Yan, H, Wu, Z, Yan, N, Wang, Z, Shi, Y. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a site-2 protease family intramembrane metalloprotease.
Science, 318, 2007
|
|
1QMV
 
 | | thioredoxin peroxidase B from red blood cells | | Descriptor: | PEROXIREDOXIN-2 | | Authors: | Isupov, M.N, Littlechild, J.A, Lebedev, A.A, Errington, N, Vagin, A.A, Schroder, E. | | Deposit date: | 1999-10-07 | | Release date: | 2000-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Decameric 2-Cys Peroxiredoxin from Human Erythrocytes at 1.7 A Resolution.
Structure, 8, 2000
|
|
5GU0
 
 | | Crystal structure of Au.CL-apo-E45C/R52C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observation of gold sub-nanocluster nucleation within a crystalline protein cage
Nat Commun, 8, 2017
|
|
3K2B
 
 | | Crystal structure of photosynthetic A4 isoform glyceraldehyde-3-phosphate dehydrogenase complexed with NAD, from Arabidopsis thaliana | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Falini, G, Thumiger, A, Sparla, F, Marri, L, Trost, P. | | Deposit date: | 2009-09-29 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase (isoform A4) from Arabidopsis thaliana in complex with NAD
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3B5D
 
 | |
1QOH
 
 | | A MUTANT SHIGA-LIKE TOXIN IIE | | Descriptor: | SHIGA-LIKE TOXIN IIE B SUBUNIT | | Authors: | Pannu, N.S, Boodhoo, A, Armstrong, G.D, Clark, C.G, Brunton, J.L, Read, R.J. | | Deposit date: | 1999-11-08 | | Release date: | 2000-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Mutant Shiga-Like Toxin Iie Bound to its Receptor Gb(3): Structure of a Group II Shiga-Like Toxin with Altered Binding Specificity
Structure, 8, 2000
|
|
5VCA
 
 | | VCP like ATPase from T. acidophilum (VAT)-Substrate bound conformation | | Descriptor: | VCP-like ATPase | | Authors: | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
5UO3
 
 | | Structure of human neuronal nitric oxide synthase heme domain in complex with 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-((methylamino)methyl)benzonitrile | | Descriptor: | 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(methylamino)methyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UK3
 
 | |
5H2K
 
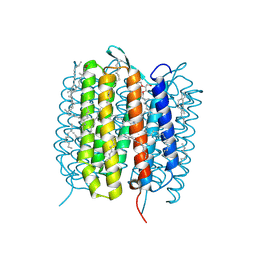 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
3B9O
 
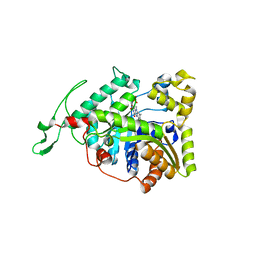 | | long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN | | Descriptor: | Alkane monooxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
3KA3
 
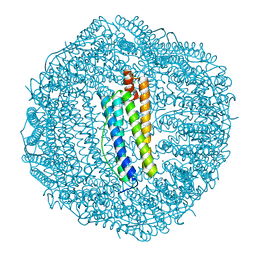 | | Frog M-ferritin with magnesium | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Tosha, T, Ng, H.L, Theil, E, Alber, T, Bhattasali, O. | | Deposit date: | 2009-10-18 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Moving Metal Ions through Ferritin-Protein Nanocages from Three-Fold Pores to Catalytic Sites.
J.Am.Chem.Soc., 132, 2010
|
|
5H3S
 
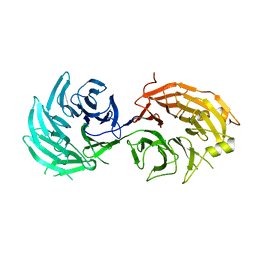 | | apo form of GEMIN5-WD | | Descriptor: | GLYCEROL, Gem-associated protein 5 | | Authors: | Bharath, S.R, Tang, X, Song, H. | | Deposit date: | 2016-10-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for specific recognition of pre-snRNA by Gemin5
Cell Res., 26, 2016
|
|
1QM9
 
 | | NMR, REPRESENTATIVE STRUCTURE | | Descriptor: | POLYPYRIMIDINE TRACT-BINDING PROTEIN | | Authors: | Conte, M.R, Grune, T, Curry, S, Matthews, S. | | Deposit date: | 1999-09-22 | | Release date: | 2000-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of Tandem RNA Recognition Motifs from Polypyrimidine Tract Binding Protein Reveals Novel Features of the Rrm Fold
Embo J., 19, 2000
|
|
5UR1
 
 | | FGFR1 kinase domain complex with SN37333 in reversible binding mode | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{1-[4-(dimethylamino)but-2-enoyl]piperidin-4-yl}-7-(phenylamino)-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1 | | Authors: | Yosaatmadja, Y, Paik, W.-K, Smaill, J.B, Squire, C.J. | | Deposit date: | 2017-02-09 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-Oxo-3, 4-dihydropyrimido[4, 5-d]pyrimidinyl derivatives as new irreversible pan fibroblast growth factor receptor (FGFR) inhibitors.
Eur J Med Chem, 135, 2017
|
|
2O2B
 
 | |
3BDY
 
 | | Dual specific bH1 Fab in complex with VEGF | | Descriptor: | Fab Fragment -Heavy Chain, Fab Fragment -Light Chain, GLYCEROL, ... | | Authors: | Bostrom, J.M, Wiesmann, C, Appleton, B.A. | | Deposit date: | 2007-11-15 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site
Science, 323, 2009
|
|
2O8Y
 
 | | Apo IRAK4 Kinase Domain | | Descriptor: | Interleukin-1 receptor-associated kinase 4 | | Authors: | Boriack-Sjodin, P.A, Mol, C. | | Deposit date: | 2006-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the apo and inhibited IRAK4 kinase domain
To be Published
|
|
2O2W
 
 | |
