6UVH
 
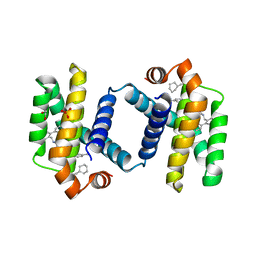 | | Crystal structure of BCL-XL bound to compound 15: (R)-2-(3-(2-((4'-Chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-(2-((4'-Chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVG
 
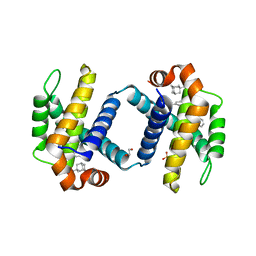 | | Crystal structure of BCL-XL bound to compound 13: (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-(((3R,5R,7R)-adamantan-1-ylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-(((3R,5R,7R)-adamantan-1-ylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6CY6
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Escherichia coli in complex with tris(hydroxymethyl)aminomethane. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-04 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1AKE
 
 | |
5R5I
 
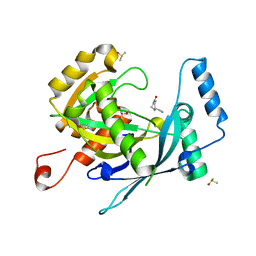 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N06325b | | Descriptor: | 1-ethanoylpiperidine-4-carbonitrile, DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6MH5
 
 | |
6D1H
 
 | | Crystal structure of NDM-1 complexed with compound 11 | | Descriptor: | ACETATE ION, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
4M8Y
 
 | | GS-8374, a Novel Phosphonate-Containing Inhibitor of HIV-1 Protease, Effectively Inhibits HIV PR Mutants with Amino Acid Insertions | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Grantz saskova, K, Brynda, J, Rezacova, P, Kozisek, M, Konvalinka, J. | | Deposit date: | 2013-08-14 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | GS-8374, a prototype phosphonate-containing inhibitor of HIV-1 protease, effectively inhibits protease mutants with amino acid insertions.
J.Virol., 88, 2014
|
|
7DXI
 
 | |
7VVD
 
 | |
2DB4
 
 | | Crystal structure of rotor ring with DCCD of the V- ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DICYCLOHEXYLUREA, SODIUM ION, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7L4G
 
 | |
5L5Y
 
 | |
1K1X
 
 | | Crystal structure of 4-alpha-glucanotransferase from thermococcus litoralis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
4ZTF
 
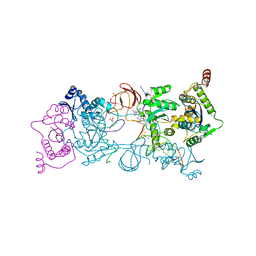 | | Crystal Structure of the Prototype Foamy Virus Intasome with a 2-Pyridinone Aminal Inhibitor | | Descriptor: | (1R,2R,5S)-8'-(3-chloro-4-fluorobenzyl)-6'-hydroxy-2'-[(2S)-2-hydroxypropyl]-9',10'-dihydro-2'H-spiro[bicyclo[3.1.0]hexane-2,3'-imidazo[5,1-a][2,6]naphthyridine]-1',5',7'(8'H)-trione, 17 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (TRANSFERRED STRAND), 19 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (NON-TRANSFERRED STRAND), ... | | Authors: | Klein, D.J, Patel, S. | | Deposit date: | 2015-05-14 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 2-Pyridinone Aminals: A Prodrug Strategy to Advance a Second Generation of HIV-1 Integrase Strand Transfer Inhibitors.
J.Med.Chem., 58, 2015
|
|
7KN1
 
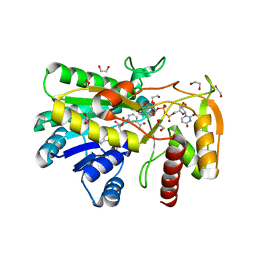 | |
5EEI
 
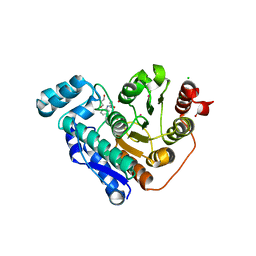 | |
7UZ3
 
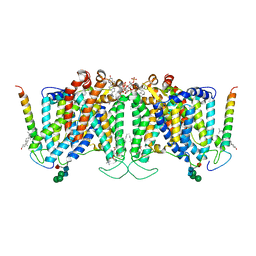 | | Band 3-Glycophorin A complex, outward facing | | Descriptor: | Band 3 anion transport protein, CHOLESTEROL, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-08 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5ZCD
 
 | | Crystal structure of Alpha-glucosidase in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
2UZC
 
 | | Structure of human PDLIM5 in complex with the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ AND LIM DOMAIN 5 | | Authors: | Bunkoczi, G, Elkins, J, Salah, E, Burgess-Brown, N, Papagrigoriou, E, Pike, A.C.W, Turnbull, A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Doyle, D. | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms.
Protein Sci., 19, 2010
|
|
4JZF
 
 | | Structure of factor VIIA in complex with the inhibitor 2-{2-[(3-carbamoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid | | Descriptor: | 2-{2-[(3-carbamoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Factor VIIa (Heavy Chain), ... | | Authors: | Wei, A, Anumula, R. | | Deposit date: | 2013-04-02 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of nonbenzamidine factor VIIa inhibitors using a biaryl acid scaffold.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2V4N
 
 | | Crystal structure of Salmonella typhimurium SurE at 1.7 angstrom resolution in orthorhombic form | | Descriptor: | GLYCEROL, MAGNESIUM ION, MULTIFUNCTIONAL PROTEIN SUR E, ... | | Authors: | Anju, P, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-09-26 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies on a Mesophilic Stationary Phase Survival Protein (Sur E) from Salmonella Typhimurium
FEBS J., 275, 2008
|
|
3L16
 
 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyridin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
3Q9O
 
 | |
5EEK
 
 | |
