5W2E
 
 | | HCV NS5B RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor MK-8876 | | Descriptor: | 2-(4-fluorophenyl)-5-(11-fluoro-6H-pyrido[2',3':5,6][1,3]oxazino[3,4-a]indol-2-yl)-N-methyl-6-[methyl(methylsulfonyl)amino]-1-benzofuran-3-carboxamide, Genome polyprotein | | Authors: | Lesburg, C.A, Ummat, A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a New Structural Class of Broadly Acting HCV Non-Nucleoside Inhibitors Leading to the Discovery of MK-8876.
ChemMedChem, 12, 2017
|
|
9FZI
 
 | | A new crystal structure of the hPXR-LBD in fusion with an SRC1 co-activator peptide and in complex with SR12813 (P212121 form) | | Descriptor: | Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1, [2-(3,5-DI-TERT-BUTYL-4-HYDROXY-PHENYL)-1-(DIETHOXY-PHOSPHORYL)-VINYL]-PHOSPHONIC ACID DIETHLYL ESTER | | Authors: | Carivenc, C, Blanc, P, Bourguet, W, Delfosse, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A two-in-one expression construct for biophysical and structural studies of the human pregnane X receptor ligand-binding domain, a pharmaceutical and environmental target.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
4ITH
 
 | | Crystal structure of RIP1 kinase in complex with necrostatin-1 analog | | Descriptor: | (5R)-5-[(7-chloro-1H-indol-3-yl)methyl]-3-methylimidazolidine-2,4-dione, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1, ... | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
5W3Z
 
 | | Crystal structure of SsoPox AsC6 mutant (L72I-Y99F-I122L-L228M-F229S-W263L) | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Hiblot, J, Gotthard, G, Jacquet, P, Daude, D, Bergonzi, C, Chabriere, E, Elias, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Rational engineering of a native hyperthermostable lactonase into a broad spectrum phosphotriesterase.
Sci Rep, 7, 2017
|
|
4D6C
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98)(L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
5AE9
 
 | | Crystal structure of mouse PI3 kinase delta in complex with GSK2292767 | | Descriptor: | N-[5-[4-(5-{[(2R,6S)-2,6-DIMETHYL-4-MORPHOLINYL]METHYL}-1,3-OXAZOL-2-YL)-1H-INDAZOL-6-YL]-2-(METHYLOXY)-3-PYRIDINYL]METHANESULFONAMIDE, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Down, K.D, Amour, A, Baldwin, I.R, Cooper, A.W.J, Deakin, A.M, Felton, L.M, Guntrip, S.B, Hardy, C, Harrison, Z.A, Jones, K.L, Jones, P, Keeling, S.E, Le, J, Livia, S, Lucas, F, Lunniss, C.J, Parr, N.J, Robinson, E, Rowland, P, Smith, S, Thomas, D.A, Vitulli, G, Washio, Y, Hamblin, N. | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Optimization of Novel Indazoles as Highly Potent and Selective Inhibitors of Phosphoinositide 3-Kinase Gamma for the Treatment of Respiratory Disease.
J.Med.Chem., 58, 2015
|
|
3QT4
 
 | | Structure of digestive procathepsin L 3 of Tenebrio molitor larval midgut | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Cathepsin-L-like midgut cysteine proteinase, PHOSPHATE ION, ... | | Authors: | Beton, D, Guzzo, C.R, Terra, W.R, Farah, C.S. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The 3D structure and function of digestive cathepsin L-like proteinases of Tenebrio molitor larval midgut.
Insect Biochem.Mol.Biol., 42, 2012
|
|
5C9M
 
 | | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-06-28 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.362 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution.
To Be Published
|
|
2V62
 
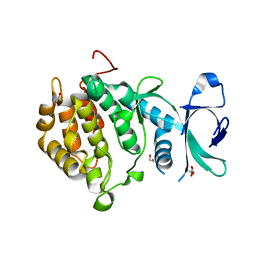 | | Structure of vaccinia-related kinase 2 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SERINE/THREONINE-PROTEIN KINASE VRK2, ... | | Authors: | Bunkoczi, G, Eswaran, J, Cooper, C, Fedorov, O, Keates, T, Rellos, P, Salah, E, Savitsky, P, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Pseudokinase Vrk3 Reveals a Degraded Catalytic Site, a Highly Conserved Kinase Fold, and a Putative Regulatory Binding Site.
Structure, 17, 2009
|
|
5TB1
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, class 1) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
6JIW
 
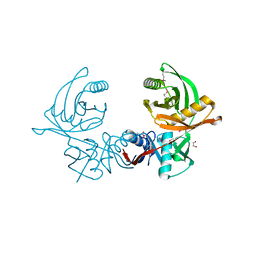 | | Crystal structure of S. aureus CntK with C72S mutation | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Luo, S, Ju, Y, Zhou, H. | | Deposit date: | 2019-02-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of CntK, the cofactor-independent histidine racemase in staphylopine-mediated metal acquisition of Staphylococcus aureus.
Int.J.Biol.Macromol., 135, 2019
|
|
3Q84
 
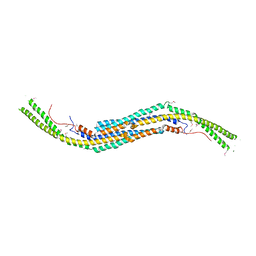 | | Crystal structure of human PACSIN 1 F-BAR domain | | Descriptor: | CALCIUM ION, Protein kinase C and casein kinase substrate in neurons protein 1 | | Authors: | Bai, X. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human PACSIN 1 F-BAR domain
To be Published
|
|
3ZQW
 
 | | Structure of CBM3b of major scaffoldin subunit ScaA from Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-12 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6GG8
 
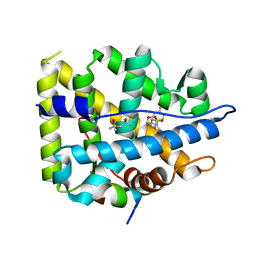 | | Mineralocorticoid receptor in complex with (s)-13 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Mineralocorticoid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Edman, K, Aagaard, A, Tangefjord, S. | | Deposit date: | 2018-05-03 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Mineralocorticoid Receptor Modulators with Low Impact on Electrolyte Homeostasis but Maintained Organ Protection.
J.Med.Chem., 62, 2019
|
|
5A8R
 
 | |
5ALI
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 1-[3-[5-(7-aminothiazolo[5,4-d]pyrimidin-2-yl)-1-[[(2S)-tetrahydrofuran-2-yl]methyl]imidazol-4-yl]phenyl]-3-(4-methoxyphenyl)urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
6D15
 
 | | Crystal structure of KPC-2 complexed with compound 1 | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, [(6,7-dimethyl-2-oxo-2H-1-benzopyran-4-yl)methyl]phosphonic acid | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
6GIZ
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - SUBSTRATE COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6D3Z
 
 | | Protease SFTI complex | | Descriptor: | Plasminogen, Trypsin inhibitor 1 | | Authors: | Law, R.H.P, Wu, G. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Highly Potent and Selective Plasmin Inhibitors Based on the Sunflower Trypsin Inhibitor-1 Scaffold Attenuate Fibrinolysis in Plasma.
J. Med. Chem., 62, 2019
|
|
6GJ2
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - COMPLEX WITH INOSITOL HEXASULPHATE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
7Y7T
 
 | | QDE-1 in complex with 12nt DNA template, ATP and 3'-dGTP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*G*AP*GP*AP*CP*CP*TP*TP*TP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
6CM1
 
 | |
2DJH
 
 | | Crystal structure of the carboxy-terminal ribonuclease domain of Colicin E5 | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, 2-AMINO-9-(2-DEOXY-3-O-PHOSPHONOPENTOFURANOSYL)-1,9-DIHYDRO-6H-PURIN-6-ONE, Colicin-E5 | | Authors: | Yajima, S, Inoue, S, Ogawa, T, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2006-04-03 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction
Nucleic Acids Res., 34, 2006
|
|
3PKZ
 
 | | Structural basis for catalytic activation of a serine recombinase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Recombinase Sin, ... | | Authors: | Keenholtz, R.A, Boocock, M.R, Rowland, S.J, Stark, W.M, Rice, P.A. | | Deposit date: | 2010-11-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for catalytic activation of a serine recombinase.
Structure, 19, 2011
|
|
5B2T
 
 | | Crystal structure of the Streptococcus pyogenes Cas9 VRER variant in complex with sgRNA and target DNA (TGCG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
