2CPI
 
 | | Solution structure of the RNA recognition motif of CNOT4 | | Descriptor: | CCR4-NOT transcription complex subunit 4 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA recognition motif of CNOT4
To be Published
|
|
7DKD
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Asn-Tyr | | Descriptor: | ASPARAGINE, Dipeptidyl-peptidase, GLYCEROL, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKC
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Tyr-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, TYROSINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKE
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Phe-Tyr | | Descriptor: | Dipeptidyl-peptidase, GLYCEROL, PHENYLALANINE, ... | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKB
 
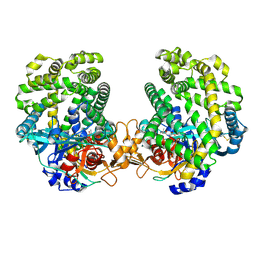 | | Stenotrophomonas maltophilia DPP7 in complex with Val-Tyr | | Descriptor: | Dipeptidyl-peptidase, TYROSINE, VALINE | | Authors: | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2020-11-23 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
3KO0
 
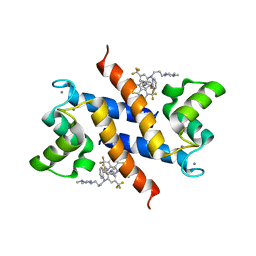 | | Structure of the tfp-ca2+-bound activated form of the s100a4 Metastasis factor | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, Protein S100-A4 | | Authors: | Malashkevich, V.N, Dulyaninova, N.G, Knight, D, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2009-11-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenothiazines inhibit S100A4 function by inducing protein oligomerization.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2MG6
 
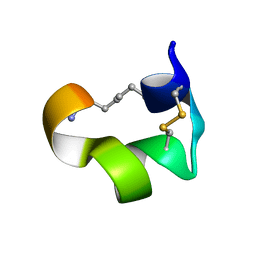 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [3,16]-trans dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-18 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
2MFY
 
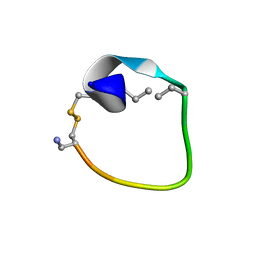 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [2,8]-trans dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2013-12-18 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
2MFX
 
 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [2,8]-cis dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2013-12-18 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
1TTL
 
 | | Omega-conotoxin GVIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
5AAN
 
 | | Crystal structure of Drosophila NCS-1 bound to penothiazine FD44 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CG5907-PA, ... | | Authors: | Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2015-07-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2Q0U
 
 | | Structure of Pectenotoxin-2 and Latrunculin B Bound to Actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | Authors: | Allingham, J.S, Miles, C.O, Rayment, I. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural basis for regulation of actin polymerization by pectenotoxins.
J.Mol.Biol., 371, 2007
|
|
2Q0R
 
 | | Structure of Pectenotoxin-2 Bound to Actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | Authors: | Allingham, J.S, Miles, C.O, Rayment, I. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural basis for regulation of actin polymerization by pectenotoxins.
J.Mol.Biol., 371, 2007
|
|
2MTO
 
 | | Non-reducible analogues of alpha-conotoxin RgIA: [2,8]-cis dicarba RgIA | | Descriptor: | Alpha-conotoxin RgIA | | Authors: | Chhabra, S, Robinson, S, Norton, R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-11-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Dicarba Analogues of alpha-Conotoxin RgIA. Structure, Stability, and Activity at Potential Pain Targets.
J.Med.Chem., 57, 2014
|
|
1FEL
 
 | | CRYSTALLOGRAPHIC STUDIES ON COMPLEXES BETWEEN RETINOIDS AND PLASMA RETINOL-BINDING PROTEIN | | Descriptor: | N-(4-HYDROXYPHENYL)ALL-TRANS RETINAMIDE, RETINOL BINDING PROTEIN | | Authors: | Zanotti, G, Marcello, M, Malpeli, G, Sartori, G, Berni, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-11-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies on complexes between retinoids and plasma retinol-binding protein.
J.Biol.Chem., 269, 1994
|
|
1FEM
 
 | | CRYSTALLOGRAPHIC STUDIES ON COMPLEXES BETWEEN RETINOIDS AND PLASMA RETINOL-BINDING PROTEIN | | Descriptor: | RETINOIC ACID, RETINOL BINDING PROTEIN | | Authors: | Zanotti, G, Marcello, M, Malpeli, G, Sartori, G, Berni, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies on complexes between retinoids and plasma retinol-binding protein.
J.Biol.Chem., 269, 1994
|
|
3V79
 
 | | Structure of human Notch1 transcription complex including CSL, RAM, ANK, and MAML-1 on HES-1 promoter DNA sequence | | Descriptor: | DNA 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', DNA 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Conformational Locking upon Cooperative Assembly of Notch Transcription Complexes.
Structure, 20, 2012
|
|
1CNL
 
 | | ALPHA-CONOTOXIN IMI | | Descriptor: | PROTEIN (ALPHA-CONOTOXIN IMI) | | Authors: | Gehrmann, J, Daly, N.L, Craik, D.J. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of alpha-conotoxin ImI by 1H nuclear magnetic resonance.
J.Med.Chem., 42, 1999
|
|
1DW4
 
 | | NMR STRUCTURE OF OMEGA-CONOTOXIN MVIIA: CONSTRAINTS ON DISULPHIDE BRIDGES | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Atkinson, R.A, Kieffer, B, Dejaegere, A, Sirockin, F, Lefevre, J.-F. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Biochemistry, 39, 2000
|
|
1DW5
 
 | | NMR STRUCTURE OF OMEGA-CONOTOXIN MVIIA: NO CONSTRAINTS ON DISULPHIDE BRIDGES | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Atkinson, R.A, Kieffer, B, Dejaegere, A, Sirockin, F, Lefevre, J.-F. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Biochemistry, 39, 2000
|
|
2B5Q
 
 | |
2B5P
 
 | |
6OVJ
 
 | |
6VY9
 
 | | Crystal structure of NotF prenyltransferase | | Descriptor: | Deoxybrevianamide E synthase notF | | Authors: | Dan, Q, Smith, J.L. | | Deposit date: | 2020-02-25 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Data Science-Driven Analysis of Substrate-Permissive Diketopiperazine Reverse Prenyltransferase NotF: Applications in Protein Engineering and Cascade Biocatalytic Synthesis of (-)-Eurotiumin A.
J.Am.Chem.Soc., 144, 2022
|
|
1GIB
 
 | | MU-CONOTOXIN GIIIB, NMR | | Descriptor: | MU-CONOTOXIN GIIIB | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-04-17 | | Release date: | 1996-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mu-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels.
Biochemistry, 35, 1996
|
|
