6XIA
 
 | |
6C1B
 
 | | FGFR1 kinase complex with inhibitor SN37118 | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{(3S)-1-[(2E)-4-(dimethylamino)but-2-enoyl]pyrrolidin-3-yl}-7-(phenylamino)-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1 | | Authors: | Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the structural requirements for covalent inhibition of FGFR1-3
To Be Published
|
|
2VLK
 
 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, Van Der Merwe, A, Bell, J, Mcmichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
3G91
 
 | | 1.2 Angstrom structure of the exonuclease III homologue Mth0212 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-12 | | Release date: | 2010-03-09 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA.
J.Mol.Biol., 399, 2010
|
|
5CYS
 
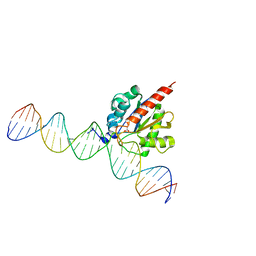 | |
4GWA
 
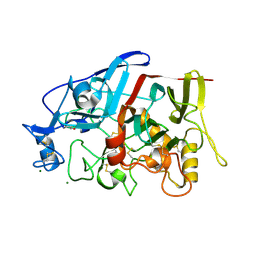 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Limnoria quadripunctata | | Descriptor: | GH7 family protein, MAGNESIUM ION | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-01 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5DHE
 
 | | Crystal structure of ChBD3 from Thermococcus kodakarensis KOD1 | | Descriptor: | Chitinase, GLYCEROL | | Authors: | Niwa, S, Hibi, M, Takeda, K, Miki, K. | | Deposit date: | 2015-08-30 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of chitin binding domains of chitinase from Thermococcus kodakarensis KOD1
Febs Lett., 590, 2016
|
|
1M2Z
 
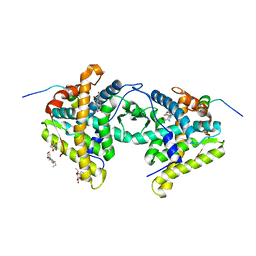 | | Crystal structure of a dimer complex of the human glucocorticoid receptor ligand-binding domain bound to dexamethasone and a TIF2 coactivator motif | | Descriptor: | DEXAMETHASONE, glucocorticoid receptor, nuclear receptor coactivator 2, ... | | Authors: | Bledsoe, R.B, Montana, V.G, Stanley, T.B, Delves, C.J, Apolito, C.J, Mckee, D.D, Consler, T.G, Parks, D.J, Stewart, E.L, Willson, T.M, Lambert, M.H, Moore, J.T, Pearce, K.H, Xu, H.E. | | Deposit date: | 2002-06-26 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Reveals a Novel Mode of Receptor Dimerization and Coactivator Recognition
Cell(Cambridge,Mass.), 110, 2002
|
|
6Y94
 
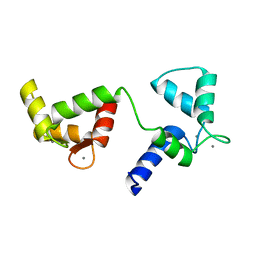 | | Ca2+-bound Calmodulin mutant N53I | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Holt, C, Nielsen, L.H, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
1M8I
 
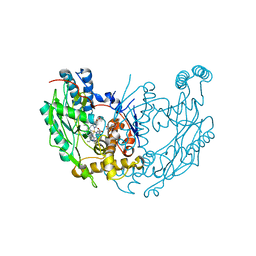 | | inducible nitric oxide synthase with 5-nitroindazole bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 5-NITROINDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Panda, K, Andersson, G, Aberg, A, Wallace, A.V, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-07-24 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Changes in Nitric Oxide Synthases Induced by Chlorzoxazone and Nitroindazoles: Crystallographic and Computational Analyses of Inhibitor Potency
Biochemistry, 41, 2002
|
|
2CN7
 
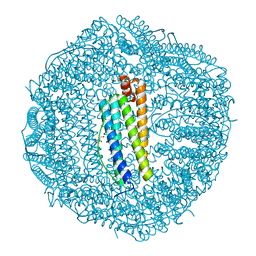 | | Recombinant human H ferritin, K86Q, E27D and E107D mutant | | Descriptor: | CALCIUM ION, FERRITIN HEAVY CHAIN, GLYCEROL | | Authors: | Toussaint, L, Crichton, R.R, Declercq, J.P. | | Deposit date: | 2006-05-18 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-Resolution X-Ray Structures of Human Apoferritin H-Chain Mutants Correlated with Their Activity and Metal-Binding Sites.
J.Mol.Biol., 365, 2007
|
|
6BGN
 
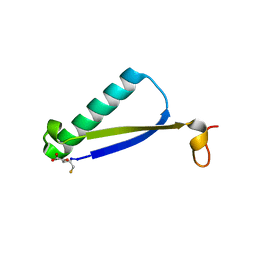 | | Crystal Structure of 4-Oxalocrotonate Tautomerase After Incubation with 5-Fluoro-2-hydroxy-2,4-pentadienoate | | Descriptor: | 2-hydroxymuconate tautomerase, 5-fluoranyl-2-oxidanylidene-pentanoic acid, GLYCEROL, ... | | Authors: | Zhang, Y, Li, W, Stack, T. | | Deposit date: | 2017-10-29 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Inactivation of 4-Oxalocrotonate Tautomerase by 5-Halo-2-hydroxy-2,4-pentadienoates.
Biochemistry, 57, 2018
|
|
5CZZ
 
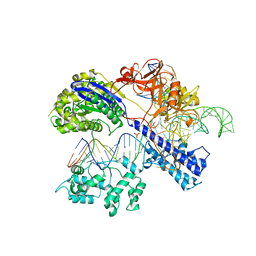 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGAAT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
1M8P
 
 | |
2VBM
 
 | | Tailspike protein of bacteriophage Sf6 complexed with tetrasaccharide | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mueller, J.J, Barbirz, S, Freiberg, A, Seckler, R, Heinemann, U. | | Deposit date: | 2007-09-14 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Intersubunit Active Site between Supercoiled Parallel Beta Helices in the Trimeric Tailspike Endorhamnosidase of Shigella Flexneri Phage Sf6.
Structure, 16, 2008
|
|
4ZK7
 
 | | Crystal structure of rescued two-component self-assembling tetrahedral cage T33-31 | | Descriptor: | Chorismate mutase, Divalent-cation tolerance protein CutA | | Authors: | Liu, Y, Cascio, D, Sawaya, M.R, Bale, J, Collazo, M.J, Park, R, King, N, Baker, D, Yeates, T. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression.
Protein Sci., 24, 2015
|
|
4GYL
 
 | | The E142L mutant of the amidase from Geobacillus pallidus showing the result of Michael addition of acrylamide at the active site cysteine | | Descriptor: | Aliphatic amidase, CHLORIDE ION, PROPIONAMIDE | | Authors: | Weber, B.W, Sewell, B.T, Kimani, S.W, Varsani, A, Cowan, D.A, Hunter, R. | | Deposit date: | 2012-09-05 | | Release date: | 2013-08-21 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
3GE8
 
 | | Toluene 4-monooxygenase HD T201A diferric, resting state complex | | Descriptor: | ACETATE ION, FE (III) ION, Toluene-4-monooxygenase system protein A, ... | | Authors: | Elsen, N.L, Bailey, L.J, Hauser, A.D, Fox, B.G. | | Deposit date: | 2009-02-25 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Role for threonine 201 in the catalytic cycle of the soluble diiron hydroxylase toluene 4-monooxygenase.
Biochemistry, 48, 2009
|
|
2CLU
 
 | | Recombinant human H ferritin, K86Q and E107D mutant | | Descriptor: | CALCIUM ION, FERRITIN HEAVY CHAIN, GLYCEROL, ... | | Authors: | Toussaint, L, Crichton, R.R, Declercq, J.P. | | Deposit date: | 2006-05-02 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Resolution X-Ray Structures of Human Apoferritin H-Chain Mutants Correlated with Their Activity and Metal-Binding Sites.
J.Mol.Biol., 365, 2007
|
|
6XM5
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-07-29 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
4GON
 
 | |
5D27
 
 | | Crystal Structure of the P-Rex1 PH domain | | Descriptor: | NICKEL (II) ION, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-08-05 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
2VFQ
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V450A | | Descriptor: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN,, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
6XMR
 
 | |
2CGN
 
 | | FACTOR INHIBITING HIF-1 ALPHA with succinate | | Descriptor: | FE (III) ION, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA INHIBITOR, SUCCINIC ACID, ... | | Authors: | McDonough, M.A, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2006-03-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Studies on the Inhibition of the Hypoxia-Inducible Transcription Factor Hydroxylases by Tricarboxylic Acid Cycle Intermediates.
J.Biol.Chem., 282, 2007
|
|
