2V9S
 
 | | Second LRR domain of human Slit2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SLIT HOMOLOG 2 PROTEIN N-PRODUCT | | Authors: | Morlot, C, Cusack, S, McCarthy, A.A. | | Deposit date: | 2007-08-25 | | Release date: | 2007-09-25 | | Last modified: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into the Slit-Robo Complex.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4WWA
 
 | | Crystal structure of binary complex Bud32-Cgi121 | | Descriptor: | EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, SULFATE ION | | Authors: | Zhang, W, van Tilbeurgh, H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
3GCF
 
 | | Terminal oxygenase of carbazole 1,9a-dioxygenase from Nocardioides aromaticivorans IC177 | | Descriptor: | CHLORIDE ION, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Inoue, K, Nojiri, H. | | Deposit date: | 2009-02-22 | | Release date: | 2009-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specific Interactions between the ferredoxin and terminal oxygenase components of a class IIB Rieske nonheme iron oxygenase, carbazole 1,9a-dioxygenase.
J.Mol.Biol., 392, 2009
|
|
5D8H
 
 | |
2VCT
 
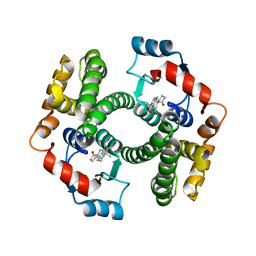 | |
2BXX
 
 | |
4H2P
 
 | | Tetrameric form of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
5D9Q
 
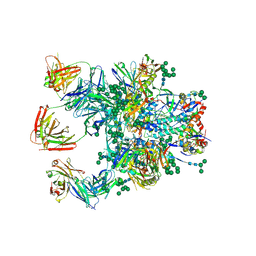 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 and scFv NIH45-46 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Ward, A.B, Wilson, I.A. | | Deposit date: | 2015-08-18 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
5DB0
 
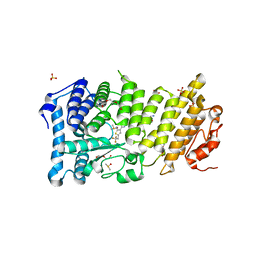 | | Menin in complex with MI-352 | | Descriptor: | 1-[(2R)-2,3-dihydroxypropyl]-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, 1-[(2S)-2,3-dihydroxypropyl]-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1 H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
2BP7
 
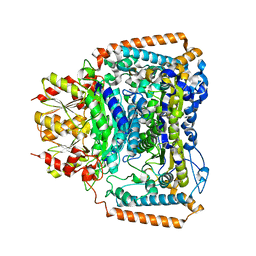 | | New crystal form of the Pseudomonas putida branched-chain dehydrogenase (E1) | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2005-04-18 | | Release date: | 2005-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Molecular Origins of Specificity in the Assembly of a Multienzyme Complex.
Structure, 13, 2005
|
|
5DBA
 
 | |
4H3Y
 
 | |
4HH0
 
 | | Dark-state structure of AppA C20S without the Cys-rich region from Rb. sphaeroides | | Descriptor: | AppA protein, CHLORIDE ION, FLAVIN MONONUCLEOTIDE | | Authors: | Winkler, A, Heintz, U, Lindner, R, Reinstein, J, Shoeman, R, Schlichting, I. | | Deposit date: | 2012-10-09 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1LYA
 
 | | CRYSTAL STRUCTURES OF NATIVE AND INHIBITED FORMS OF HUMAN CATHEPSIN D: IMPLICATIONS FOR LYSOSOMAL TARGETING AND DRUG DESIGN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATHEPSIN D, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Baldwin, E.T, Bhat, T.N, Gulnik, S, Erickson, J.W. | | Deposit date: | 1993-04-22 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
6XKN
 
 | |
4Z0D
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038Trp] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
6XN3
 
 | | Structure of the Lactococcus lactis Csm CTR_4:3 CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
5DD9
 
 | | Menin in complex with MI-326 | | Descriptor: | 4-[4-(5-methyl-1,3,4-thiadiazol-2-yl)piperazin-1-yl]-6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidine, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rational Design of Orthogonal Multipolar Interactions with Fluorine in Protein-Ligand Complexes.
J.Med.Chem., 58, 2015
|
|
2CCO
 
 | |
6XN7
 
 | | Structure of the Lactococcus lactis Csm NTR CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
6XJY
 
 | |
2VDC
 
 | | THE 9.5 A RESOLUTION STRUCTURE OF GLUTAMATE SYNTHASE FROM CRYO-ELECTRON MICROSCOPY AND ITS OLIGOMERIZATION BEHAVIOR IN SOLUTION: FUNCTIONAL IMPLICATIONS. | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Cottevieille, M, Larquet, E, Jonic, S, Petoukhov, M.V, Caprini, G, Paravisi, S, Svergun, D.I, Vanoni, M.A, Boisset, N. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-Mda Hexamer by Cryoelectron Microscopy and its Oligomerization Behavior in Solution: Functional Implications.
J.Biol.Chem., 283, 2008
|
|
6XJQ
 
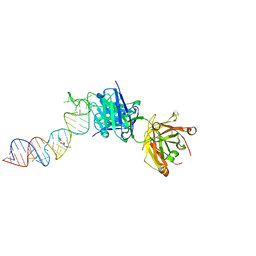 | | Crystal structure of a self-alkylating ribozyme - alkylated form with biotinylated epoxide substrate | | Descriptor: | 2-{[(4R)-4-hydroxyhexyl]oxy}ethyl 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoate, Fab HAVx Heavy Chain, Fab HAVx Light Chain, ... | | Authors: | Koirala, D, Piccirilli, J.A. | | Deposit date: | 2020-06-24 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Structural basis for substrate binding and catalysis by a self-alkylating ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
6XJZ
 
 | |
3G8M
 
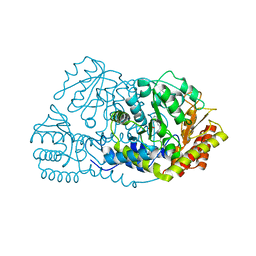 | | Serine Hydroxymethyltransferase Y55F Mutant | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase | | Authors: | Angelucci, F, Ilari, A. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Role of a conserved active site cation-pi interaction in Escherichia coli serine hydroxymethyltransferase.
Biochemistry, 48, 2009
|
|
