9BJ1
 
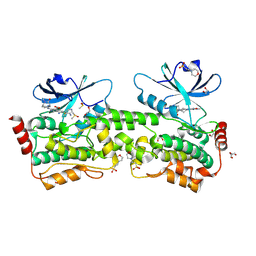 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (4S,5R,7R,11aP)-10-{[(3R)-3-hydroxy-1-methyl-2-oxopyrrolidin-3-yl]ethynyl}-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, (9S)-2-{[(6P)-8-amino-6-(5-amino-4-methylpyridin-3-yl)-7-fluoroisoquinolin-3-yl]amino}-6-methyl-5,6-dihydro-4H-pyrazolo[1,5-d][1,4]diazepin-7(8H)-one, 1,2-ETHANEDIOL, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
9C4P
 
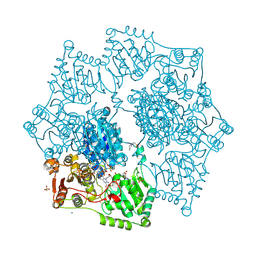 | | Crystal structure of wild-type arabidopsis thaliana acetohydroxyacid synthase in complex with commercial herbicide triasulfuron | | Descriptor: | 2-(2-chloroethoxy)-N-[(4-methoxy-6-methyl-1,3,5-triazin-2-yl)carbamoyl]benzene-1-sulfonamide, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cheng, Y, Guddat, L.W. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Acetohydroxyacid Synthase in Complex with the Herbicide Triasulfuron and Two Analogues with Herbicidal Activity in Field Trials.
J.Agric.Food Chem., 2024
|
|
8R4Y
 
 | | Homotrimer for the lumenal domain of Vacuolar Sorting Receptor 1 (VSR1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Vacuolar-sorting receptor 1, alpha-D-mannopyranose, ... | | Authors: | Borges, R.J, Eldahshoury, M.K, Nettleship, J, Bird, L, An, J, Levada, A.J.L, Shah, N, Thomson, M, Fontes, M.R.M, Owens, R, Denecke, J, Postis, V, Uson, I, Goldman, A, De Marcos Lousa, C. | | Deposit date: | 2023-11-14 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The crystal structure of Vacuolar Sorting Receptor 1 (VSR1) lumenal domain reveals a stable domain-swapped trimer and a potential new cargo binding site
To Be Published
|
|
6R1O
 
 | | Crystal structure of E. coli seryl-tRNA synthetase complexed to a seryl sulfamoyl adenosine derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
4PNL
 
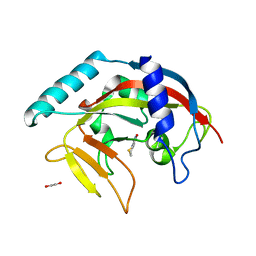 | | Crystal structure of TNKS-2 in complex with DR2313. | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,7,8-tetrahydro-4H-thiopyrano[4,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5WH5
 
 | |
5DT2
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD11 [N4-methyl-N2-(2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl)pyrimidine-2,4-diamine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~4~-methyl-N~2~-[2-methyl-1-(2-phenoxyphenyl)-1H-indol-6-yl]pyrimidine-2,4-diamine, ... | | Authors: | Scheufler, C, Gaul, C, Be, C, Moebitz, H. | | Deposit date: | 2015-09-17 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Novel Dot1L Inhibitors through a Structure-Based Fragmentation Approach.
Acs Med.Chem.Lett., 7, 2016
|
|
7BHR
 
 | | Crystal structure of MAT2a with triazinone fragment 1 bound in the allosteric site | | Descriptor: | 4-(dimethylamino)-6-ethoxy-1~{H}-1,3,5-triazin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
6DMN
 
 | |
3P5P
 
 | | Crystal Structure of Taxadiene Synthase from Pacific Yew (Taxus brevifolia) in complex with Mg2+ and 13-aza-13,14-dihydrocopalyl diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-(methyl{2-[(1S,4aS,8aS)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]ethyl}amino)ethyl trihydrogen diphosphate, MAGNESIUM ION, ... | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2010-10-10 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis.
Nature, 469, 2011
|
|
6DNB
 
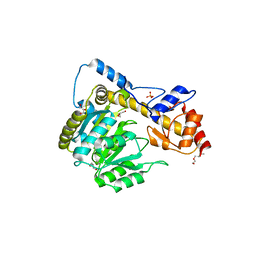 | | Crystal structure of T110A:S256A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, GLYCEROL, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
8TQ7
 
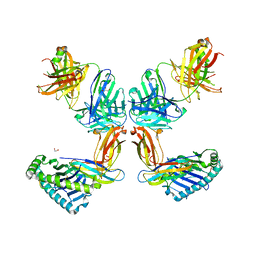 | | Crystal structure of Fab.34.2.12 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab 34.2.12 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
5I23
 
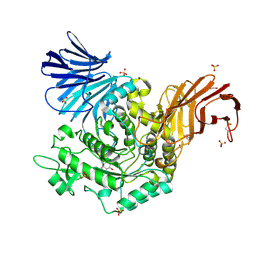 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with Cyclophellitol Aziridine probe CF022 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-{[(1S,2R,3S,4S,5R,6R)-2,3,4,5-tetrahydroxy-6-(hydroxymethyl)cyclohexyl]amino}octyl)triaza-1,2-dien-2-ium, Oligosaccharide 4-alpha-D-glucosyltransferase, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2016-02-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detection of Active Mammalian GH31 alpha-Glucosidases in Health and Disease Using In-Class, Broad-Spectrum Activity-Based Probes.
Acs Cent.Sci., 2, 2016
|
|
6V1O
 
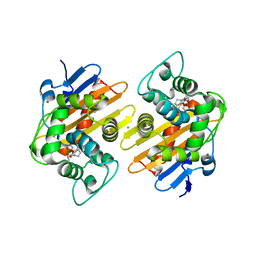 | | Structure of OXA-48 bound to QPX7728 at 1.80 A | | Descriptor: | (1aR,7bS)-5-fluoro-2-hydroxy-1,1a,2,7b-tetrahydrocyclopropa[c][1,2]benzoxaborinine-4-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Boronic Acid QPX7728, an Ultrabroad-Spectrum Inhibitor of Serine and Metallo-beta-lactamases.
J.Med.Chem., 63, 2020
|
|
7V76
 
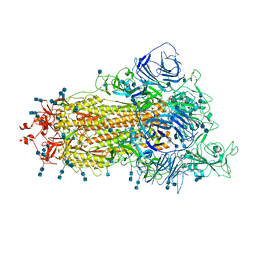 | | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), uncleavable form, one RBD-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), uncleavable form, one RBD-up conformation
To Be Published
|
|
7V7Z
 
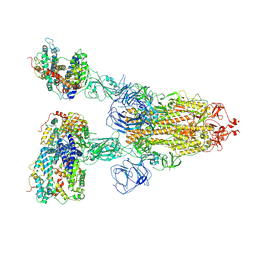 | | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form
To Be Published
|
|
7V86
 
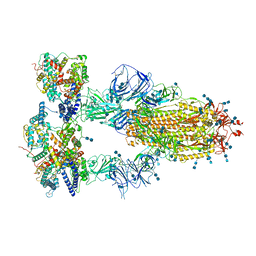 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form
To Be Published
|
|
7V8A
 
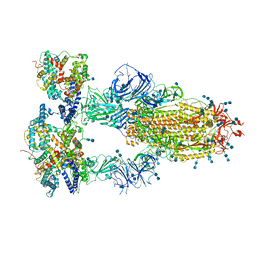 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2
To Be Published
|
|
7V8C
 
 | | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), Cleavable form, one RBD-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), Cleavable form, one RBD-up conformation
To Be Published
|
|
6HDG
 
 | | D170N variant of beta-phosphoglucomutase from Lactococcus lactis complexed with beta-G1P in a closed conformer to 1.2 A. | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Wood, H.P, Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
8OIP
 
 | | 28S mammalian mitochondrial small ribosomal subunit with mtRF1 and P-site tRNA | | Descriptor: | 12S rRNA, 28S ribosomal protein S15, mitochondrial, ... | | Authors: | Saurer, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Ban, N. | | Deposit date: | 2023-03-23 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of translation termination at noncanonical stop codons in human mitochondria.
Science, 380, 2023
|
|
7V82
 
 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 1
To Be Published
|
|
7V7U
 
 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), two RBD-up conformation 2
To Be Published
|
|
7V7R
 
 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 4
To Be Published
|
|
8H6J
 
 | | Cryo-EM structure of human exon-defined spliceosome in the mature pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
