9IK9
 
 | | Cryo-EM Structure of SST analogs bond SSTR1-Gi complex | | Descriptor: | (4J2)(DCY)(DTY)(DTR)K(DVA)(DCY)(ALO)(NH2), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Du, Y. | | Deposit date: | 2024-06-26 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into the binding modes of lanreotide and pasireotide with somatostatin receptor 1.
Acta Pharm Sin B, 15, 2025
|
|
5YLN
 
 | |
8T6F
 
 | | Crystal structure of human MBP-Myeloid cell leukemia 1 (Mcl-1) in complex with BRD810 inhibitor | | Descriptor: | (3aM,9S,15R)-4-chloro-3-ethyl-7-{3-[(6-fluoronaphthalen-1-yl)oxy]propyl}-2-methyl-15-[2-(morpholin-4-yl)ethyl]-2,10,11,12,13,15-hexahydropyrazolo[4',3':9,10][1,6]oxazacycloundecino[8,7,6-hi]indole-8-carboxylic acid, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Poncet-Montange, G, Lemke, C.T. | | Deposit date: | 2023-06-15 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | BRD-810 is a highly selective MCL1 inhibitor with optimized in vivo clearance and robust efficacy in solid and hematological tumor models.
Nat Cancer, 5, 2024
|
|
7XAO
 
 | | Crystal structure of thioredoxin 1 | | Descriptor: | Thioredoxin | | Authors: | Chang, Y.J, Park, H.H. | | Deposit date: | 2022-03-18 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structure of Acinetobacter baumannii thioredoxin 1.
Biochem.Biophys.Res.Commun., 608, 2022
|
|
9INP
 
 | | Human Pin1 (Peptidyl-prolyl cis-trans isomerase) catalytic domain in complex with a covalent inhibitor | | Descriptor: | CITRIC ACID, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ~{N}-[(3~{R})-1,1-bis(oxidanylidene)thiolan-3-yl]-2-chloranyl-~{N}-(2,2-dimethylpropyl)-5-nitro-pyrimidin-4-amine | | Authors: | Wang, X.Y, Tian, M.Z, Zhou, J, Xu, B.L. | | Deposit date: | 2024-07-08 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of Novel Pyrimidine Derivatives as Human Pin1 Covalent Inhibitors.
Acs Med.Chem.Lett., 16, 2025
|
|
9F2H
 
 | | Crystal structure of SARS-CoV-2 N-protein C-terminal domain in complex with riluzole | | Descriptor: | 1,2-ETHANEDIOL, 6-(trifluoromethoxy)-1,3-benzothiazol-2-amine, Nucleoprotein, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B, Perez-Canadillas, J.M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The ALS drug riluzole binds to the C-terminal domain of SARS-CoV-2 nucleocapsid protein and has antiviral activity.
Structure, 33, 2025
|
|
3LXH
 
 | | Crystal Structure of Cytochrome P450 CYP101D1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Cytochrome P450, PHOSPHATE ION, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
4HSG
 
 | | Crystal structure of human PRMT3 in complex with an allosteric inhibitor (PRMT3- KTD) | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-(2-oxo-2-phenylethyl)urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dobrovetsky, E, Dong, A, Liu, F, Li, F, Tempel, W, Siarheyeva, A, Hajian, T, Smil, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting an allosteric binding site of PRMT3 yields potent and selective inhibitors.
J. Med. Chem., 56, 2013
|
|
5YL8
 
 | | The crystal structure of inactive dimeric peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.79 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase, SODIUM ION | | Authors: | Bairagya, H.R, Sharma, P, Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The crystal structure of inactive dimeric peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.79 A resolution
To Be Published
|
|
4HTK
 
 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 2.17 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7CI8
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | (1S)-1-[1-[(5-phenyl-1,2-oxazol-3-yl)methyl]imidazol-2-yl]ethanol, MAGNESIUM ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mima, M, Baker, L.M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
8XW7
 
 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate and ADP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Nucleotide Selectivity in Pyruvate Kinase.
J.Mol.Biol., 436, 2024
|
|
7XH0
 
 | | crystal structure of Csn-PD from Paenibacillus dendritiformis | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Chitosanase | | Authors: | Sun, H.H, Cheng, Y.M, Cao, R, Liu, Q, Zhao, L. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | crystal structure of Csn-PD from Paenibacillus dendritiformis
To Be Published
|
|
1JV7
 
 | | BACTERIORHODOPSIN O-LIKE INTERMEDIATE STATE OF THE D85S MUTANT AT 2.25 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
9I7K
 
 | |
4Y8D
 
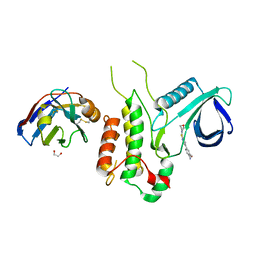 | | Crystal structure of Cyclin-G associated kinase (GAK) complexed with selective 12i inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-[3-(morpholin-4-yl)[1,2]thiazolo[4,3-b]pyridin-6-yl]aniline, Cyclin-G-associated kinase, ... | | Authors: | Chaikuad, A, Heroven, C, Nowak, R, De Jonghe, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents.
J.Med.Chem., 58, 2015
|
|
6UK6
 
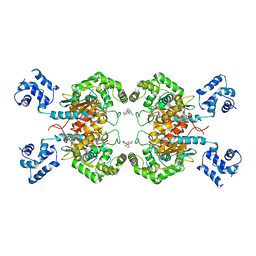 | |
8K3J
 
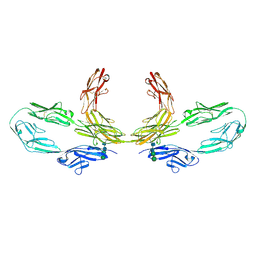 | | Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2, ... | | Authors: | Zhang, Z.Z. | | Deposit date: | 2023-07-16 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer
To Be Published
|
|
6RFE
 
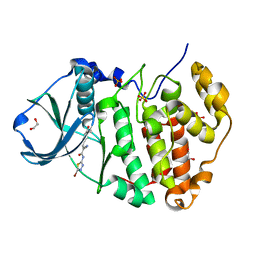 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 4 | | Descriptor: | (~{E})-~{N}-(5-bromanyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | Deposit date: | 2019-04-13 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
4YD4
 
 | |
4YEK
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with thymidine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
9IK8
 
 | | Cryo-EM Structure of SSTR1-Gi SST analogs complex | | Descriptor: | DTR-LYS-TY5-PHA-A1D5E-004, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Du, Y. | | Deposit date: | 2024-06-26 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural insights into the binding modes of lanreotide and pasireotide with somatostatin receptor 1.
Acta Pharm Sin B, 15, 2025
|
|
5YIG
 
 | | Crystal structure of Streptococcus pneumonia ParE with inhibitor | | Descriptor: | 1-ethyl-3-[5-[2-[(1S,5R)-3-methyl-3,8-diazabicyclo[3.2.1]octan-8-yl]-5-(2-oxidanylidene-3H-1,3,4-oxadiazol-5-yl)pyridin-3-yl]-4-[4-(trifluoromethyl)-1,3-thiazol-2-yl]pyridin-2-yl]urea, DNA topoisomerase 4 subunit B | | Authors: | Cherian, J, Tan, Y, Hill, J. | | Deposit date: | 2017-10-04 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of dual GyrB/ParE inhibitors active against Gram-negative bacteria.
Eur J Med Chem, 157, 2018
|
|
7MLC
 
 | | PYL10 bound to the ABA pan-antagonist 4a | | Descriptor: | 1-{2-[3,5-dicyclopropyl-4-(4-{[(quinoxaline-2-carbonyl)amino]methyl}-1H-1,2,3-triazol-1-yl)phenyl]acetamido}cyclohexane-1-carboxylic acid, Abscisic acid receptor PYL10, GLYCEROL | | Authors: | Peterson, F.C, Vaidya, A.S, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2021-04-28 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Click-to-lead design of a picomolar ABA receptor antagonist with potent activity in vivo.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6BQ9
 
 | | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida | | Descriptor: | CHLORIDE ION, DNA topoisomerase 4 subunit A, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida.
To Be Published
|
|
