6WG1
 
 | |
3G43
 
 | |
5CR5
 
 | |
4IQH
 
 | |
6BAL
 
 | | 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CHLORIDE ION, Malate dehydrogenase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate
To Be Published
|
|
2EQ8
 
 | |
4WZA
 
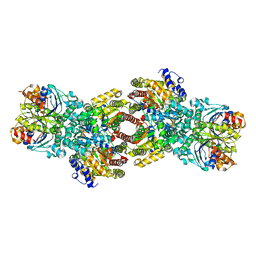 | | Asymmetric Nucleotide Binding in the Nitrogenase Complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-11-19 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8995 Å) | | Cite: | Structural evidence for asymmetrical nucleotide interactions in nitrogenase.
J.Am.Chem.Soc., 137, 2015
|
|
2DZJ
 
 | | 2DZJ/Solution Structure of the N-terminal Ubiquitin-like Domain in Human Synaptic Glycoprotein SC2 | | Descriptor: | Synaptic glycoprotein SC2 | | Authors: | Zhao, C, Yoneyama, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-29 | | Release date: | 2007-03-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 2DZJ/Solution Structure of the N-terminal Ubiquitin-like Domain in Human Synaptic Glycoprotein SC2
To be Published
|
|
4X33
 
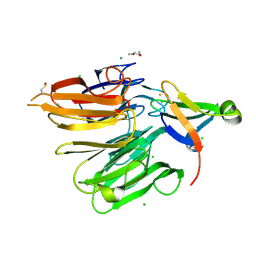 | | Structure of the Elongator cofactor complex Kti11/Kti13 at 1.45A | | Descriptor: | 1,2-DIMETHOXYETHANE, CHLORIDE ION, Diphthamide biosynthesis protein 3, ... | | Authors: | Kolaj-Robin, O, McEwen, A.G, Cavarelli, J, Seraphin, B. | | Deposit date: | 2014-11-27 | | Release date: | 2015-01-21 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Elongator cofactor complex Kti11/Kti13 provides insight into the role of Kti13 in Elongator-dependent tRNA modification.
Febs J., 282, 2015
|
|
2X86
 
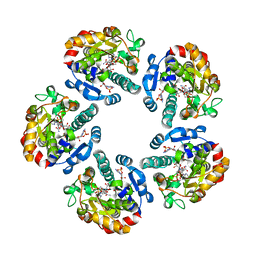 | | AGME bound to ADP-B-mannose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-L-GLYCERO-D-MANNO-HEPTOSE-6-EPIMERASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kowatz, T, Morrison, J.P, Tanner, M.E, Naismith, J.H. | | Deposit date: | 2010-03-06 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Y140F Mutant of Adp-L-Glycero-D-Manno-Heptose 6-Epimerase Bound to Adp-Beta-D-Mannose Suggests a One Base Mechanism.
Protein Sci., 19, 2010
|
|
2WK2
 
 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitotrio-thiazoline dithioamide. | | Descriptor: | 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose-(1-4)-2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CHITINASE A, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-04 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3G6J
 
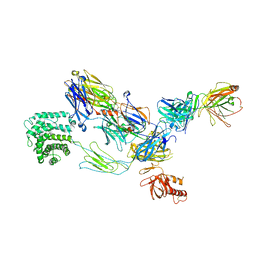 | | C3b in complex with a C3b specific Fab | | Descriptor: | CALCIUM ION, Complement C3 alpha chain, Complement C3 beta chain, ... | | Authors: | Wiesmann, C. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Functional Analysis of a C3b-specific Antibody That Selectively Inhibits the Alternative Pathway of Complement
J.Biol.Chem., 284, 2009
|
|
1LFA
 
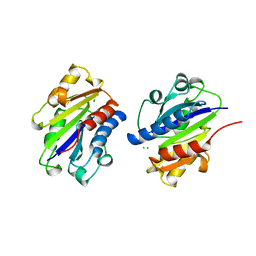 | | CD11A I-DOMAIN WITH BOUND MN++ | | Descriptor: | CD11A, CHLORIDE ION, MANGANESE (II) ION | | Authors: | Leahy, D.J, Qu, A. | | Deposit date: | 1995-09-08 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, alpha L beta 2) integrin.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2WRT
 
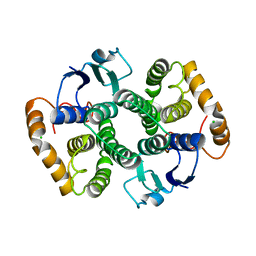 | | The 2.4 Angstrom structure of the Fasciola hepatica mu class GST, GST26 | | Descriptor: | CHLORIDE ION, GLUTATHIONE S-TRANSFERASE CLASS-MU 26 KDA ISOZYME 51 | | Authors: | Line, K, Isupov, M.N, LaCourse, E.J, Brophy, P.M, Littlechild, J.A. | | Deposit date: | 2009-09-02 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.5 Angstrom Structure of a Mu Class Gst from Fasciola Hepatica
To be Published
|
|
4IJI
 
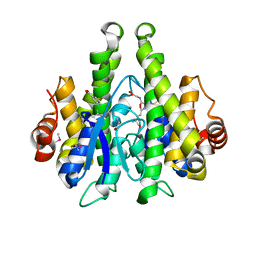 | | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione | | Descriptor: | ACRYLIC ACID, BENZOIC ACID, Glutathione S-transferase-like protein YibF, ... | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione
To be Published
|
|
2X56
 
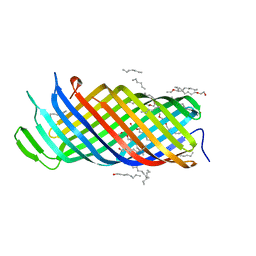 | | Yersinia Pestis Plasminogen Activator Pla (Native) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, COAGULASE/FIBRINOLYSIN | | Authors: | Eren, E, Murphy, M, Goguen, J, van den Berg, B. | | Deposit date: | 2010-02-05 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Active Site Water Network in the Plasminogen Activator Pla from Yersinia Pestis
Structure, 18, 2010
|
|
2WUC
 
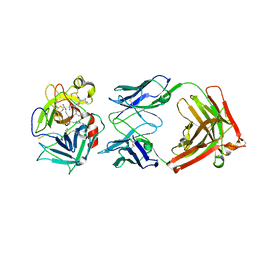 | | Crystal structure of HGFA in complex with the allosteric non- inhibitory antibody Fab40.deltaTrp and Ac-KQLR-chloromethylketone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-KQLR-CHLOROMETHYLKETONE INHIBITOR, FAB FRAGMENT FAB40.DELTATRP HEAVY CHAIN, ... | | Authors: | Ganesan, R, Eigenbrot, C, Shia, S. | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unraveling the Allosteric Mechanism of Serine Protease Inhibition by an Antibody
Structure, 17, 2009
|
|
5CSO
 
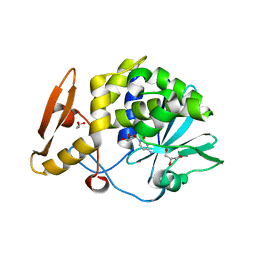 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleoside, cytidine at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
6WC7
 
 | |
6WDV
 
 | |
3GAH
 
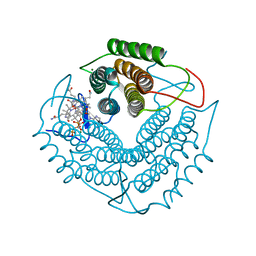 | | Structure of a F112H variant PduO-type ATP:corrinoid adenosyltransferase from Lactobacillus reuteri complexed with cobalamin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, Cobalamin adenosyltransferase PduO-like protein, ... | | Authors: | St Maurice, M, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2009-02-17 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Residue Phe112 of the human-type corrinoid adenosyltransferase (PduO) enzyme of Lactobacillus reuteri is critical to the formation of the four-coordinate Co(II) corrinoid substrate and to the activity of the enzyme.
Biochemistry, 48, 2009
|
|
2ENY
 
 | | Solution structure of the ig-like domain (2735-2825) of human obscurin | | Descriptor: | Obscurin | | Authors: | Nagashima, K, Izumi, K, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ig-like domain (2735-2825) of human obscurin
To be Published
|
|
3GC9
 
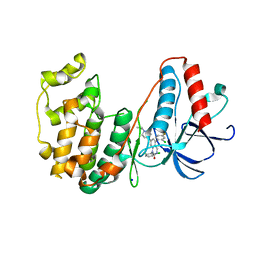 | | The structure of p38beta C119S, C162S in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 5-(2-chloro-4-fluorophenyl)-1-(2,6-dichlorophenyl)-7-[1-(1-methylethyl)piperidin-4-yl]-3,4-dihydroquinazolin-2(1H)-one, Mitogen-activated protein kinase 11, SODIUM ION, ... | | Authors: | Scapin, G, Patel, S.B. | | Deposit date: | 2009-02-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The three-dimensional structure of MAP kinase p38beta: different features of the ATP-binding site in p38beta compared with p38alpha.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4X5L
 
 | | Crystal structure of Dscam1 Ig7 domain, isoform 9 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AM, SODIUM ION | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
5CST
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine diphosphate at 1.78 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
