8TK2
 
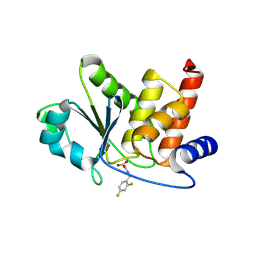 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) complexed with 2-((2,4-difluorobenzyl)amino)-2-oxoacetic acid | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, Dual specificity protein phosphatase 3, ... | | Authors: | Aleshin, A.E, Wu, J, Lambert, L.J, Cosford, N.D.P, Tautz, L. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) binding 2-((2,4-difluorobenzyl)amino)-2-oxoacetic acid
To Be Published
|
|
5W9S
 
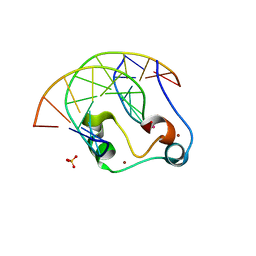 | | Zinc finger of human CXXC5 in complex with CpG DNA | | Descriptor: | CXXC-type zinc finger protein 5, CpG DNA fragment, SULFATE ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
8TQT
 
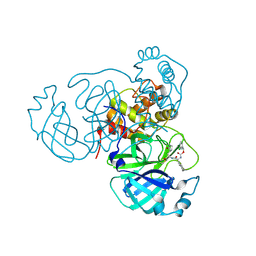 | | MPI52 bound to Mpro of SARS-CoV-2 | | Descriptor: | (3-chlorophenyl)methyl [(2S)-3-cyclohexyl-1-({(1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | MPI52 bound to SARS-CoV-2 Mpro
To Be Published
|
|
8TQL
 
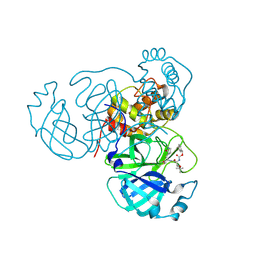 | | MPI54 bound to Mpro of SARS-CoV-2 | | Descriptor: | 3C-like proteinase nsp5, benzyl [(2S,3S)-3-tert-butoxy-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxobutan-2-yl]carbamate | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MPI54 bound to SARS-CoV-2 Mpro
To Be Published
|
|
6DMN
 
 | |
6DUB
 
 | | Crystal structure of a methyltransferase | | Descriptor: | Alpha N-terminal protein methyltransferase 1B, GLYCEROL, RCC1, ... | | Authors: | Dong, C, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2.
Commun Biol, 1, 2018
|
|
5FH6
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with compound 10 | | Descriptor: | (3~{R})-3-(piperidin-1-ylmethyl)-2,3-dihydro-1~{H}-pyrrolo[1,2-a]quinazolin-5-one, Protein polybromo-1 | | Authors: | Tallant, C, Sutherell, C.L, Siejka, P, Krojer, T, Picaud, S, Fonseca, M, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Ley, S.V, Knapp, S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Development of 2,3-Dihydropyrrolo[1,2-a]quinazolin-5(1H)-one Inhibitors Targeting Bromodomains within the Switch/Sucrose Nonfermenting Complex.
J.Med.Chem., 59, 2016
|
|
8TK6
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) in apo form | | Descriptor: | Dual specificity protein phosphatase 3 | | Authors: | Aleshin, A.E, Wu, J, Lambert, L.J, Cosford, N.D.P, Tautz, L. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragment Screening Platform and Discovery of Novel Fragment Binders of the VHR Phosphatase, a Drug Target for Sepsis and Septic Shock
To Be Published
|
|
5WBV
 
 | | Crystal Structure of the SET Domain of Human SUV420H1 In Complex With Inhibitor | | Descriptor: | 2-chloro-5-(4-methyl-6-oxo-3-phenylpyrano[2,3-c]pyrazol-1(6H)-yl)benzoic acid, Histone-lysine N-methyltransferase KMT5B, S-ADENOSYLMETHIONINE, ... | | Authors: | Halabelian, L, Tempel, W, Brown, P.J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-29 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the SET Domain of Human SUV420H1 In Complex With Inhibitor
To be published
|
|
5WA9
 
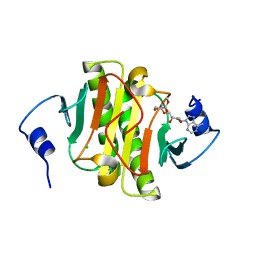 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Ala phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~{R})-1-methoxy-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2017-06-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5FAJ
 
 | |
6DNJ
 
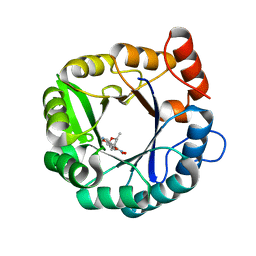 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 28 round 5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-nitro-1,2-benzoxazole, Kemp eliminase KE07 | | Authors: | Jackson, C.J, Hong, N.-S, Carr, P.D. | | Deposit date: | 2018-06-06 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
5WC9
 
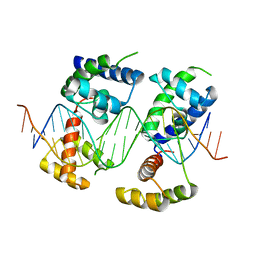 | | Human Pit-1 and 4xCATT DNA complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*TP*CP*AP*TP*TP*CP*AP*TP*TP*CP*AP*TP*TP*CP*GP*GP*A)-3'), DNA (5'-D(*CP*CP*GP*AP*AP*TP*GP*AP*AP*TP*GP*AP*AP*TP*GP*AP*AP*TP*GP*GP*T)-3'), Pituitary-specific positive transcription factor 1 | | Authors: | Agarwal, S, Cho, T.Y. | | Deposit date: | 2017-06-29 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Biochemical and structural characterization of a novel cooperative binding mode by Pit-1 with CATT repeats in the macrophage migration inhibitory factor promoter.
Nucleic Acids Res., 46, 2018
|
|
5FHF
 
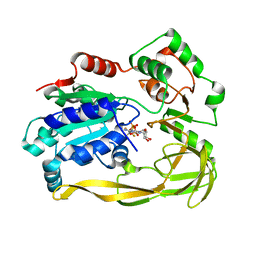 | | Crystal structure of Bacteroides sp Pif1 in complex with ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TETRAFLUOROALUMINATE ION, Uncharacterized protein | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5WCP
 
 | | Phosphotriesterase variant S7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-07-01 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphotriesterase variant S7
To Be Published
|
|
6K3G
 
 | | Crystal structure of 10-Hydroxygeraniol Dehydrogenase from Cantharanthus roseus in complex with NADP+ | | Descriptor: | 10-hydroxygeraniol oxidoreductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Sandholu, A.S, Sharmila, P.M, Thulasiram, H.V, Kulkarni, K.A. | | Deposit date: | 2019-05-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural studies on 10-hydroxygeraniol dehydrogenase: A novel linear substrate-specific dehydrogenase from Catharanthus roseus.
Proteins, 88, 2020
|
|
5WDG
 
 | | Acetolactate Synthase from Klebsiella pneumoniae in Complex with a Reaction Intermediate | | Descriptor: | (2S,3S)-2,3-dihydroxy-3-[(7S,8R,9aS)-8-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2,7-dimethyl-5,7,8,10-tetrahydro-9aH-pyrimido[4,5-d][1,3]thiazolo[3,2-a]pyrimidin-9a-yl]-2-methylbutanoic acid, Acetolactate synthase, catabolic, ... | | Authors: | Latta, A.J, Andrews, F.H, McLeish, M.J. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Acetolactate Synthase from Klebsiella pneumoniae in Complex with Mechanism-Based Inhibitor
To Be Published
|
|
6DOW
 
 | |
5WEE
 
 | | Crystal structure of HpVAL4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2017-07-09 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Heligmosomoides polygyrus Venom Allergen-like Protein-4 (HpVAL-4) is a sterol binding protein.
Int. J. Parasitol., 48, 2018
|
|
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
6DP9
 
 | |
5WEP
 
 | | Crystal structure of fosfomycin resistance protein FosA3 with inhibitor (ANY1) bound | | Descriptor: | 6,6'-(4-nitro-1H-pyrazole-3,5-diyl)bis(3-bromopyrazolo[1,5-a]pyrimidin-2(1H)-one), FosA3, ZINC ION | | Authors: | Klontz, E.H, Sundberg, E.J. | | Deposit date: | 2017-07-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Small-Molecule Inhibitor of FosA Expands Fosfomycin Activity to Multidrug-Resistant Gram-Negative Pathogens.
Antimicrob. Agents Chemother., 63, 2019
|
|
6KMN
 
 | | Crystal Structure of Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Deferrochelatase/peroxidase EfeB, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
6DPM
 
 | |
5FCZ
 
 | |
