5EPZ
 
 | |
8FX7
 
 | | Non-ribosomal PCP-C didomain (ester stabilised leucine) acceptor bound state | | Descriptor: | 2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl L-leucinate, PCP-C didomain | | Authors: | Ho, Y.T.C, Cryle, M.J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Not always an innocent bystander: the impact of stabilised phosphopantetheine moieties when studying nonribosomal peptide biosynthesis.
Chem.Commun.(Camb.), 59, 2023
|
|
8GJ4
 
 | |
8GKM
 
 | |
8GLC
 
 | |
8GIC
 
 | |
8GJP
 
 | |
2A46
 
 | |
2A48
 
 | | Crystal structure of amFP486 E150Q | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like fluorescent chromoprotein amFP486 | | Authors: | Henderson, J.N, Remington, S.J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5EHB
 
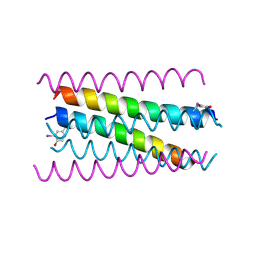 | | A de novo designed hexameric coiled-coil peptide with iodotyrosine | | Descriptor: | pHiosYI | | Authors: | Lizatovic, R, Aurelius, O, Stenstrom, O, Drakenberg, T, Akke, M, Logan, D.T, Andre, I. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-15 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | A De Novo Designed Coiled-Coil Peptide with a Reversible pH-Induced Oligomerization Switch.
Structure, 24, 2016
|
|
5EQO
 
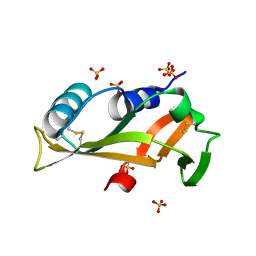 | |
5FL5
 
 | | Three dimensional structure of human carbonic anhydrase IX in complex with 5-(1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)thiophene-2- sulfonamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[1-(4-methoxyphenyl)-1,2,3-triazol-4-yl]thiophene-2-sulfonamide, CARBONIC ANHYDRASE IX, ... | | Authors: | Leitans, J, Tars, K, Zalubovskis, R. | | Deposit date: | 2015-10-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An Efficient Expression and Crystallization System of the Cancer Asociated Carbonic Anhydrase Isoform Ix.
J.Med.Chem., 58, 2015
|
|
8Q19
 
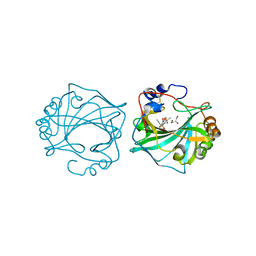 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with Sulfonamide | | Descriptor: | 2-but-2-ynyl-1,1,3-tris(oxidanylidene)-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
8Q18
 
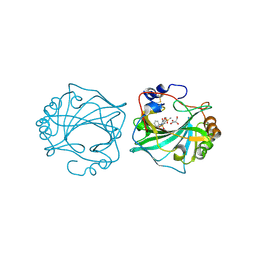 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with Sulfonamide | | Descriptor: | 1,1,3-tris(oxidanylidene)-2-(2-phenylethyl)-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
8Q1A
 
 | | The Crystal Structure of Human Carbonic Anhydrase IX in Complex with inhibitor | | Descriptor: | 1,1,3-tris(oxidanylidene)-2-pentyl-1,2-benzothiazole-6-sulfonamide, Carbonic anhydrase 9, GLYCEROL, ... | | Authors: | Leitans, J, Tars, K. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Saccharin Derivative Inhibition of Carbonic Anhydrase IX.
Chemmedchem, 18, 2023
|
|
1HK8
 
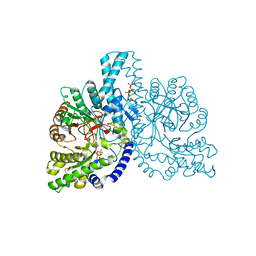 | | STRUCTURAL BASIS FOR ALLOSTERIC SUBSTRATE SPECIFICITY REGULATION IN CLASS III RIBONUCLEOTIDE REDUCTASES: NRDD IN COMPLEX WITH DGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE, MANGANESE (II) ION, ... | | Authors: | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | Deposit date: | 2003-03-06 | | Release date: | 2003-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Metal-Binding Site in the Catalytic Subunit of Anaerobic Ribonucleotide Reductase.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1F77
 
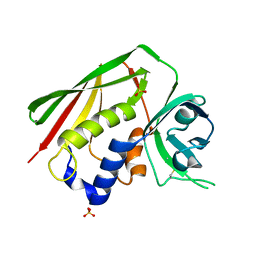 | | STAPHYLOCOCCAL ENTEROTOXIN H DETERMINED TO 2.4 A RESOLUTION | | Descriptor: | ENTEROTOXIN H, SULFATE ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
2QC5
 
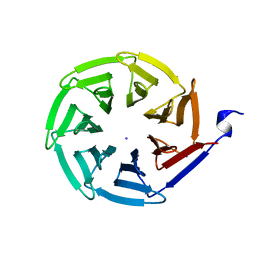 | | Streptogramin B lyase structure | | Descriptor: | IODIDE ION, Streptogramin B lactonase | | Authors: | Lipka, M, Bochtler, M. | | Deposit date: | 2007-06-19 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mechanism of the Staphylococcus cohnii virginiamycin B lyase (Vgb).
Biochemistry, 47, 2008
|
|
2AUM
 
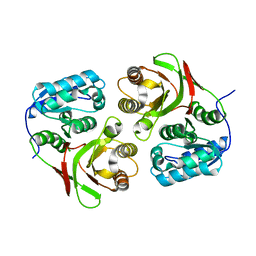 | |
8CGQ
 
 | |
8C67
 
 | | Crystal structure of Ab25 Fab | | Descriptor: | antibody 25 heavy chain, antibody 25 light chain | | Authors: | Nyblom, M, Izadi, A, Tang, D, Bahnan, W, Happonen, L, Malmstroem, J, Shannon, O, Malmstroem, L, Nordenfelt, P. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Engineering of IgG1 hinge to the flexible IgG3 hinge enhances immune defense against streptococci
To Be Published
|
|
8CPL
 
 | |
8CRG
 
 | | E. coli adenylate kinase in complex with two ADP molecules as a result of enzymatic AP4A hydrolysis | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase | | Authors: | Oelker, M, Tischlik, S, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Insights into Enzymatic Catalysis from Binding and Hydrolysis of Diadenosine Tetraphosphate by E. coli Adenylate Kinase.
Biochemistry, 62, 2023
|
|
6L05
 
 | | Crystal structure of uPA_H99Y in complex with 50F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-6-(1-benzofuran-2-yl)-Ncarbamimidoyl-pyrazine-2-carboxamide, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of uPA_H99Y in complex with 50F
To Be Published
|
|
8F0L
 
 | |
