3H1R
 
 | | Order-disorder structure of fluorescent protein FP480 | | 分子名称: | Fluorescent protein FP480 | | 著者 | Pletnev, S, Morozova, K.S, Verkhusha, V.V, Dauter, Z. | | 登録日 | 2009-04-13 | | 公開日 | 2009-09-08 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Rotational order-disorder structure of fluorescent protein FP480
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3GWH
 
 | | Crystallographic Ab Initio protein solution far below atomic resolution | | 分子名称: | PHOSPHATE ION, Transcriptional antiterminator (BglG family) | | 著者 | Rodriguez, D.D, Grosse, C, Himmel, S, Gonzalez, C, Becker, S, Sheldrick, G.M, Uson, I. | | 登録日 | 2009-04-01 | | 公開日 | 2010-04-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystallographic ab initio protein structure solution below atomic resolution
Nat.Methods, 6, 2009
|
|
3H1Z
 
 | | Molecular basis for the association of PIPKIgamma -p90 with the clathrin adaptor AP-2 | | 分子名称: | AP-2 complex subunit beta-1, Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma | | 著者 | Vahedi-Faridi, A, Kahlfeldt, N, Schaefer, J.G, Krainer, G, Keller, S, Saenger, W, Krauss, M, Haucke, V. | | 登録日 | 2009-04-14 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Molecular basis for association of PIPKI gamma-p90 with clathrin adaptor AP-2.
J.Biol.Chem., 285, 2010
|
|
3GXI
 
 | | Crystal structure of acid-beta-glucosidase at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | 著者 | Lieberman, R.L. | | 登録日 | 2009-04-02 | | 公開日 | 2009-05-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
3GZF
 
 | | Structure of the C-terminal domain of nsp4 from Feline Coronavirus | | 分子名称: | Replicase polyprotein 1ab, SULFATE ION | | 著者 | Manolaridis, I, Wojdyla, J.A, Panjikar, S, Snijder, E.J, Gorbalenya, A.E, Coutard, B, Tucker, P.A. | | 登録日 | 2009-04-07 | | 公開日 | 2009-08-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.756 Å) | | 主引用文献 | Structure of the C-terminal domain of nsp4 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3H3D
 
 | |
3H3I
 
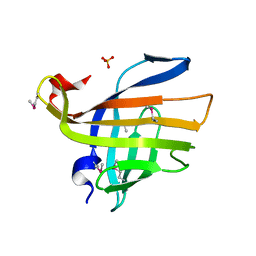 | |
3H5I
 
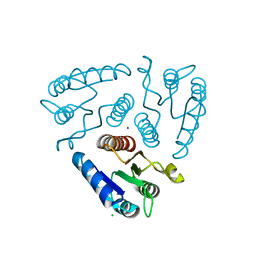 | | Crystal structure of the N-terminal domain of a response regulator/sensory box/GGDEF 3-domain protein from Carboxydothermus hydrogenoformans | | 分子名称: | CHLORIDE ION, Response regulator/sensory box protein/GGDEF domain protein, SODIUM ION | | 著者 | Bonanno, J.B, Gilmore, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-04-22 | | 公開日 | 2009-05-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the N-terminal domain of a response regulator/sensory box/GGDEF 3-domain protein from Carboxydothermus hydrogenoformans
To be Published
|
|
3H3O
 
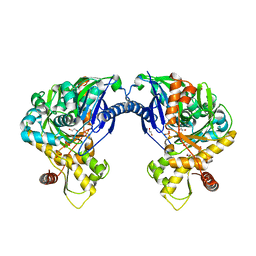 | |
3H5R
 
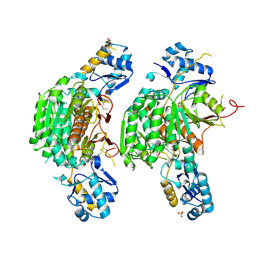 | | Crystal structure of E. coli MccB + Succinimide | | 分子名称: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | 著者 | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | 登録日 | 2009-04-22 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H6W
 
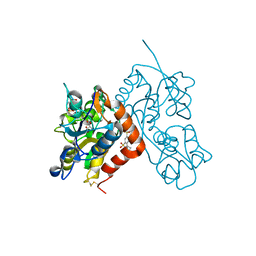 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5217 at 1.50 A resolution | | 分子名称: | (3R)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3H50
 
 | |
3H7C
 
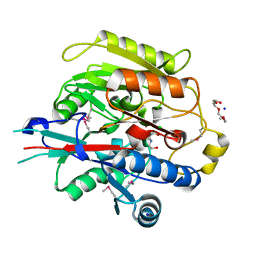 | | Crystal Structure of Arabidopsis thaliana Agmatine Deiminase from Cell Free Expression | | 分子名称: | 2,2',2''-NITRILOTRIETHANOL, Agmatine deiminase, CHLORIDE ION, ... | | 著者 | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2009-04-24 | | 公開日 | 2009-05-26 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism of Arabidopsis thaliana Agmatine Deiminase
To be Published
|
|
3H81
 
 | |
3H7S
 
 | | Crystal structures of K63-linked di- and tri-ubiquitin reveal a highly extended chain architecture | | 分子名称: | Ubiquitin, ZINC ION | | 著者 | Weeks, S.D, Grasty, K.C, Hernandez-Cuebas, L, Loll, P.J. | | 登録日 | 2009-04-28 | | 公開日 | 2009-09-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of Lys-63-linked tri- and di-ubiquitin reveal a highly extended chain architecture.
Proteins, 77, 2009
|
|
3GKM
 
 | |
3GLX
 
 | | Crystal Structure Analysis of the DtxR(E175K) complexed with Ni(II) | | 分子名称: | Diphtheria toxin repressor, NICKEL (II) ION, PHOSPHATE ION | | 著者 | D'Aquino, J.A, Denninger, A, Moulin, A, D'Aquino, K.E, Ringe, D. | | 登録日 | 2009-03-12 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Decreased sensitivity to changes in the concentration of metal ions as the basis for the hyperactivity of DtxR(E175K).
J.Mol.Biol., 390, 2009
|
|
3GMB
 
 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase | | 分子名称: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | 登録日 | 2009-03-13 | | 公開日 | 2009-04-14 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
3HC5
 
 | | FXR with SRC1 and GSK826 | | 分子名称: | 3-(6-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-1-benzothiophen-2-yl)benzoic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | 著者 | Williams, S.P, Madauss, K.P. | | 登録日 | 2009-05-05 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | FXR agonist activity of conformationally constrained analogs of GW 4064.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HCE
 
 | |
3GNF
 
 | | P1 Crystal structure of the N-terminal R1-R7 of murine MVP | | 分子名称: | Major vault protein | | 著者 | Querol-Audi, J, Casanas, A, Uson, I, Luque, D, Caston, J.R, Fita, I, Verdaguer, N. | | 登録日 | 2009-03-17 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Embo J., 28, 2009
|
|
3GWR
 
 | |
3GX0
 
 | | Crystal Structure of GSH-dependent Disulfide bond Oxidoreductase | | 分子名称: | GST-like protein yfcG, OXIDIZED GLUTATHIONE DISULFIDE | | 著者 | Ladner, J.E, Harp, J.M, Wadington, M.C, Armstrong, R.N. | | 登録日 | 2009-04-01 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Analysis of the structure and function of YfcG from Escherichia coli reveals an efficient and unique disulfide bond reductase.
Biochemistry, 48, 2009
|
|
3GO8
 
 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC3-loop deletion complex | | 分子名称: | 5'-D(*GP*CP*GP*TP*CP*CP*(8OG)P*GP*AP*TP*CP*TP*AP*C)-3', 5'-D(P*GP*GP*TP*AP*GP*AP*TP*CP*CP*GP*GP*AP*CP*G)-3', Formamidopyrimidine-DNA glycosylase, ... | | 著者 | Spong, M.C, Qi, Y, Verdine, G.L. | | 登録日 | 2009-03-18 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme
Nature, 462, 2009
|
|
3GXH
 
 | |
