3GFC
 
 | | Crystal Structure of Histone-binding protein RBBP4 | | 分子名称: | Histone-binding protein RBBP4 | | 著者 | Amaya, M.F, Dong, A, Li, Z, He, H, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | 登録日 | 2009-02-26 | | 公開日 | 2009-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
3GFM
 
 | |
3GG8
 
 | | Crystal structure of the Toxoplasma gondii Pyruvate Kinase N terminal truncated | | 分子名称: | GLYCEROL, Pyruvate kinase, SULFATE ION | | 著者 | Wernimont, A.K, Lew, J, Allali-Hassani, A, Vedadi, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hills, T, Schapira, M, Hui, R, Pizarro, J.C, Structural Genomics Consortium (SGC) | | 登録日 | 2009-02-27 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | The crystal structure of Toxoplasma gondii pyruvate kinase 1.
Plos One, 5, 2010
|
|
3GIS
 
 | |
3GCQ
 
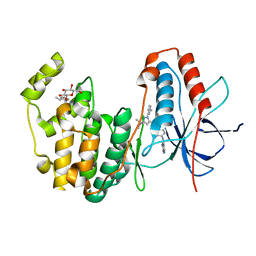 | | Human P38 MAP kinase in complex with RL45 | | 分子名称: | 1-{4-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | 著者 | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | 登録日 | 2009-02-22 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors.
J.Am.Chem.Soc., 131, 2009
|
|
3GDP
 
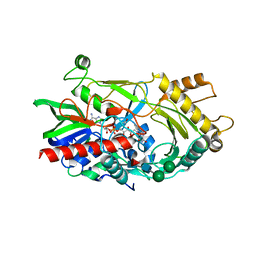 | | Hydroxynitrile lyase from almond, monoclinic crystal form | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Dreveny, I, Gruber, K, Kratky, C. | | 登録日 | 2009-02-24 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
Biochemistry, 48, 2009
|
|
3GF3
 
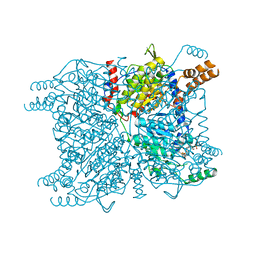 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaconyl-coA | | 分子名称: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | 著者 | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | 登録日 | 2009-02-26 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GG3
 
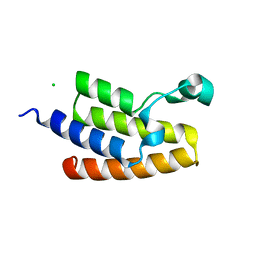 | | Crystal Structure of the Bromodomain of Human PCAF | | 分子名称: | CHLORIDE ION, Histone acetyltransferase PCAF | | 著者 | Filippakopoulos, P, Keates, T, Picaud, S, Rehana, K, Fedorov, O, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2009-02-27 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3GK2
 
 | | X-ray structure of bovine SBi279,Ca(2+)-S100B | | 分子名称: | (Z)-2-[2-(4-methylpiperazin-1-yl)benzyl]diazenecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | 著者 | Charpentier, T.H, Weber, D.J, Toth, E.A. | | 登録日 | 2009-03-09 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.984 Å) | | 主引用文献 | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3GK4
 
 | | X-ray structure of bovine SBi523,Ca(2+)-S100B | | 分子名称: | CALCIUM ION, Protein S100-B, ethyl 5-{[(1R)-1-(ethoxycarbonyl)-2-oxopropyl]sulfanyl}-1,2-dihydro[1,2,3]triazolo[1,5-a]quinazoline-3-carboxylate | | 著者 | Charpentier, T.H, Weber, D.J, Toth, E.A. | | 登録日 | 2009-03-09 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3GKA
 
 | |
3GLC
 
 | |
3GK1
 
 | | X-ray structure of bovine SBi132,Ca(2+)-S100B | | 分子名称: | 2-[(5-hex-1-yn-1-ylfuran-2-yl)carbonyl]-N-methylhydrazinecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | 著者 | Charpentier, T.H, Weber, D.J, Toth, E.A. | | 登録日 | 2009-03-09 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3GKF
 
 | |
3GMA
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaryl-CoA | | 分子名称: | Glutaconyl-CoA decarboxylase subunit A, glutaryl-coenzyme A | | 著者 | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | 登録日 | 2009-03-13 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GPG
 
 | |
3GPP
 
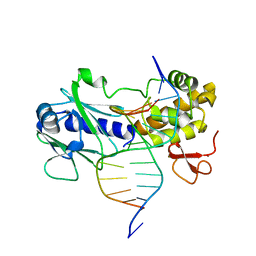 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC3-T224P complex | | 分子名称: | DNA (5'-D(P*CP*GP*TP*CP*CP*(8OG)P*GP*AP*TP*CP*TP*AP*C)-3'), DNA (5'-D(P*GP*GP*TP*AP*GP*AP*TP*CP*CP*GP*GP*AP*C)-3'), DNA glycosylase, ... | | 著者 | Spong, M.C, Qi, Y, Verdine, G.L. | | 登録日 | 2009-03-23 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
3GN6
 
 | |
3GPY
 
 | | Sequence-matched MutM Lesion Recognition Complex 3 (LRC3) | | 分子名称: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*TP*CP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*T*GP*CP*GP*TP*CP*CP*(8OG)P*GP*AP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | 著者 | Qi, Y, Verdine, G.L. | | 登録日 | 2009-03-23 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
3GNQ
 
 | |
3GPI
 
 | | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus | | 分子名称: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase | | 著者 | Ramagopal, U.A, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-03-23 | | 公開日 | 2009-04-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus
To be published
|
|
3H4Q
 
 | |
3H4W
 
 | |
3H7K
 
 | |
3H8O
 
 | | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked host-guest system | | 分子名称: | 5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*(MA7)P*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*TP*AP*TP*AP*AP*TP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | 著者 | Lu, L, Yi, C, Jian, X, Zheng, Q. | | 登録日 | 2009-04-29 | | 公開日 | 2010-03-31 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system.
Nucleic Acids Res., 38, 2010
|
|
