8OYV
 
 | |
8OYX
 
 | |
8P1B
 
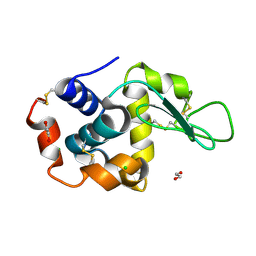 | | Lysozyme structure solved from serial crystallography data collected at 2 kHz with JUNGFRAU detector at MAXIV | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | 著者 | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | 登録日 | 2023-05-11 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8OYY
 
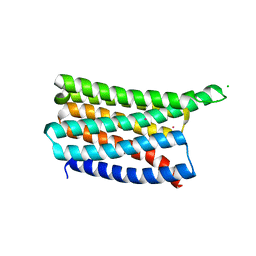 | |
8P1C
 
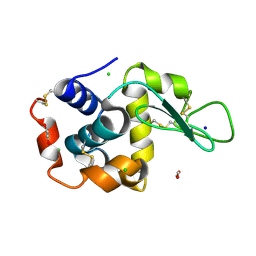 | | Lysozyme structure solved from serial crystallography data collected at 1 kHz with JUNGFRAU detector at MAXIV | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | 登録日 | 2023-05-11 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8OYW
 
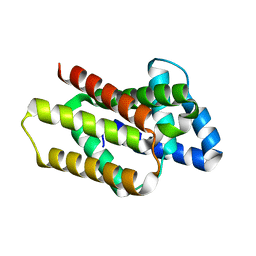 | |
7U51
 
 | |
8OJN
 
 | |
8P1A
 
 | | Lysozyme structure solved from serial crystallography data collected at 2 kHz for 5 seconds with JUNGFRAU detector at MAXIV | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | 著者 | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | 登録日 | 2023-05-11 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8P1D
 
 | | Lysozyme structure solved from serial crystallography data collected at 100 Hz with JUNGFRAU detector at MAXIV | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | 登録日 | 2023-05-11 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
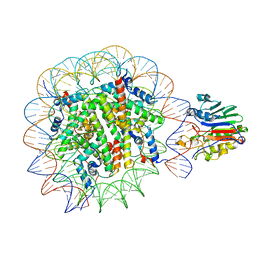
7U50
 
 | |
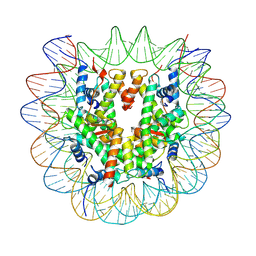
7U52
 
 | |
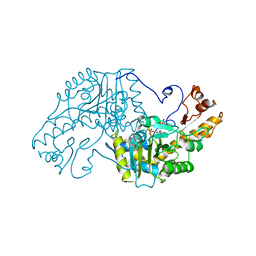
3AAT
 
 | |
3ADK
 
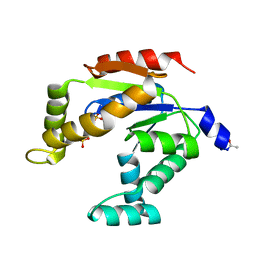 | |
7EJO
 
 | | Structure of ERH-2 bound to TOST-1 | | 分子名称: | Enhancer of rudimentary homolog 2, Enhancer of rudimentary homolog 2,Protein tost-1 | | 著者 | Wang, X, Xu, C. | | 登録日 | 2021-04-02 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.191 Å) | | 主引用文献 | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
2H1V
 
 | | Crystal structure of the Lys87Ala mutant variant of Bacillus subtilis ferrochelatase | | 分子名称: | Ferrochelatase, MAGNESIUM ION | | 著者 | Hansson, M.D, Karlberg, T, Arys Rahardja, M, Al-Karadaghi, S, Hansson, M. | | 登録日 | 2006-05-17 | | 公開日 | 2007-01-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Amino Acid Residues His183 and Glu264 in Bacillus subtilis Ferrochelatase Direct and Facilitate the Insertion of Metal Ion into Protoporphyrin IX
Biochemistry, 46, 2007
|
|
1REJ
 
 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 1 | | 分子名称: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-HYDROXYBENZOATE, cAMP-dependent protein kinase, alpha-catalytic subunit | | 著者 | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | 登録日 | 2003-11-06 | | 公開日 | 2004-02-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
5O3X
 
 | | Structural characterization of the fast and promiscuous macrocyclase from plant - apo PCY1 | | 分子名称: | CACODYLATE ION, Peptide cyclase 1 | | 著者 | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | 登録日 | 2017-05-25 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
1W60
 
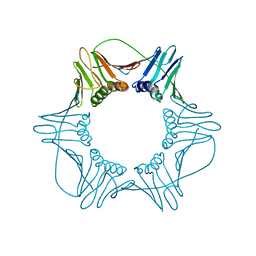 | | NATIVE HUMAN PCNA | | 分子名称: | PROLIFERATING CELL NUCLEAR ANTIGEN | | 著者 | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M. | | 登録日 | 2004-08-11 | | 公開日 | 2005-01-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4AUO
 
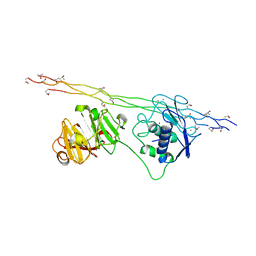 | | Crystal structure of MMP-1(E200A) in complex with a triple-helical collagen peptide | | 分子名称: | CALCIUM ION, INTERSTITIAL COLLAGENASE, TRIPLE-HELICAL COLLAGEN PEPTIDE, ... | | 著者 | Manka, S.W, Carafoli, F, Visse, R, Bihan, D, Raynal, N, Farndale, R.W, Murphy, G, Enghild, J.J, Hohenester, E, Nagase, H. | | 登録日 | 2012-05-18 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Insights Into Triple-Helical Collagen Cleavage by Matrix Metalloproteinase 1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8PW8
 
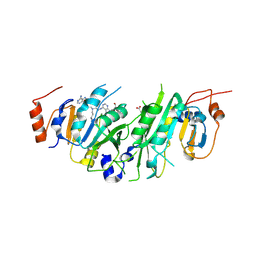 | | Crystal structure of the human METTL3-METTL14 in complex with a bisubstrate analogue (BA2) | | 分子名称: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[2-[[9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-6-yl]amino]ethyl]amino]-2-azanyl-butanoic acid, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | 著者 | Bedi, R.K, Etheve-Quelquejeu, M, Caflisch, A. | | 登録日 | 2023-07-19 | | 公開日 | 2023-11-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The catalytic mechanism of the RNA methyltransferase METTL3.
Elife, 12, 2024
|
|
1REK
 
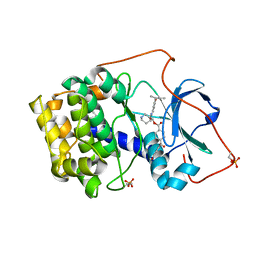 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 8 | | 分子名称: | 3-[(3-SEC-BUTYL-4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-(2-HYDROXY-5-METHOXYBENZOYL)BENZOATE, PENTANAL, cAMP-dependent protein kinase, ... | | 著者 | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | 登録日 | 2003-11-06 | | 公開日 | 2004-02-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
8PF9
 
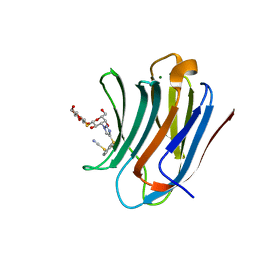 | | Galectin-3C in complex with a triazolesulfane derivative | | 分子名称: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[oxidanyl(phenyl)-$l^{3}-sulfanyl]-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | 著者 | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | 登録日 | 2023-06-15 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8PBF
 
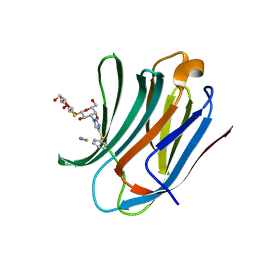 | | Galectin-3C in complex with a triazolesulfane derivative | | 分子名称: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-(4-phenylsulfanyl-1,2,3-triazol-1-yl)oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, THIOCYANATE ION | | 著者 | Kumar, R, Mahanti, M. | | 登録日 | 2023-06-09 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
1R9U
 
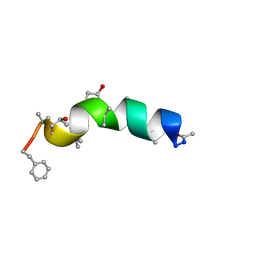 | | Refined structure of peptaibol zervamicin IIB in methanol solution from trans-hydrogen bond J couplings | | 分子名称: | ZERVAMICIN IIB | | 著者 | Shenkarev, Z.O, Balashova, T.A, Yakimenko, Z.A, Ovchinnikova, T.V, Arseniev, A.S. | | 登録日 | 2003-10-31 | | 公開日 | 2004-11-09 | | 最終更新日 | 2018-10-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Biosynthetic Uniform 13C,15N-Labelling of Zervamicin Iib. Complete 13C and 15N NMR Assignment.
J.Pept.Sci., 9, 2003
|
|
