7K51
 
 | | Mid-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure II) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K50
 
 | | Pre-translocation non-frameshifting(CCA-A) complex (Structure I) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K54
 
 | | Mid-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure II-FS) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K52
 
 | | Near post-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure III) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K55
 
 | | Near post-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure III-FS) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
3EIP
 
 | | CRYSTAL STRUCTURE OF COLICIN E3 IMMUNITY PROTEIN: AN INHIBITOR TO A RIBOSOME-INACTIVATING RNASE | | 分子名称: | PROTEIN (COLICIN E3 IMMUNITY PROTEIN), ZINC ION | | 著者 | Li, C, Zhao, D, Djebli, A, Shoham, M. | | 登録日 | 1999-03-29 | | 公開日 | 1999-11-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of colicin E3 immunity protein: an inhibitor of a ribosome-inactivating RNase.
Structure Fold.Des., 7, 1999
|
|
3V14
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein complexed with Trehalose at 1.70 A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | 著者 | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2011-12-09 | | 公開日 | 2012-01-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the complex of type I Ribosome inactivating protein complexed with Trehalose at 1.70 A resolution
To be Published
|
|
7QH6
 
 | | Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 1 | | 分子名称: | 16S ribosomal RNA, 39S ribosomal protein L13, mitochondrial, ... | | 著者 | Rebelo-Guiomar, P, Pellegrino, S, Dent, K.C, Warren, A.J, Minczuk, M. | | 登録日 | 2021-12-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | A late-stage assembly checkpoint of the human mitochondrial ribosome large subunit.
Nat Commun, 13, 2022
|
|
6ZU5
 
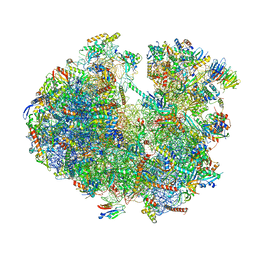 | | Structure of the Paranosema locustae ribosome in complex with Lso2 | | 分子名称: | 18S rRNA, 25S rRNA, 5S rRNA, ... | | 著者 | Ehrenbolger, K, Jespersen, N, Sharma, H, Sokolova, Y.Y, Tokarev, Y.S, Vossbrinck, C.R, Barandun, J. | | 登録日 | 2020-07-21 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Differences in structure and hibernation mechanism highlight diversification of the microsporidian ribosome.
Plos Biol., 18, 2020
|
|
4JYA
 
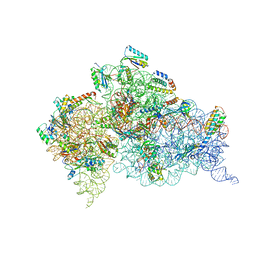 | | Crystal structures of pseudouridinilated stop codons with ASLs | | 分子名称: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | 登録日 | 2013-03-29 | | 公開日 | 2013-06-26 | | 最終更新日 | 2013-08-21 | | 実験手法 | X-RAY DIFFRACTION (3.098 Å) | | 主引用文献 | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4JV5
 
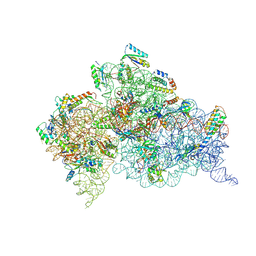 | | Crystal structures of pseudouridinilated stop codons with ASLs | | 分子名称: | 16S ribosomal RNA, 30S ribosomal protein 20, 30S ribosomal protein S10, ... | | 著者 | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | 登録日 | 2013-03-25 | | 公開日 | 2013-06-26 | | 最終更新日 | 2013-08-21 | | 実験手法 | X-RAY DIFFRACTION (3.162 Å) | | 主引用文献 | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
1DGZ
 
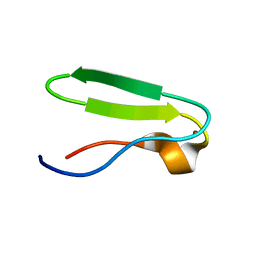 | | RIBOSMAL PROTEIN L36 FROM THERMUS THERMOPHILUS: NMR STRUCTURE ENSEMBLE | | 分子名称: | PROTEIN (L36 RIBOSOMAL PROTEIN), ZINC ION | | 著者 | Hard, T, Rak, A, Allard, P, Kloo, L, Garber, M. | | 登録日 | 1999-11-27 | | 公開日 | 1999-12-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold.
J.Mol.Biol., 296, 2000
|
|
1MI6
 
 | | Docking of the modified RF2 X-ray structure into the Low Resolution Cryo-EM map of RF2 E.coli 70S Ribosome | | 分子名称: | peptide chain release factor RF-2 | | 著者 | Rawat, U.B.S, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | 登録日 | 2002-08-22 | | 公開日 | 2003-01-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12.8 Å) | | 主引用文献 | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
1PC8
 
 | | Crystal Structure of a novel form of mistletoe lectin from Himalayan Viscum album L. at 3.8A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Himalayan mistletoe ribosome-inactivating protein, ... | | 著者 | Mishra, V, Ethayathulla, A.S, Paramasivam, M, Singh, G, Yadav, S, Kaur, P, Sharma, R.S, Babu, C.R, Singh, T.P. | | 登録日 | 2003-05-16 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of a novel ribosome-inactivating protein from a hemi-parasitic plant inhabiting the northwestern Himalayas.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4V7F
 
 | | Arx1 pre-60S particle. | | 分子名称: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | 登録日 | 2013-12-10 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (8.7 Å) | | 主引用文献 | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
4UTQ
 
 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | 分子名称: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | 著者 | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | 登録日 | 2014-07-22 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
3J5L
 
 | | Structure of the E. coli 50S subunit with ErmBL nascent chain | | 分子名称: | 23S ribsomal RNA, 5'-R(*CP*(MA6))-3', 5'-R(*CP*CP*A)-3', ... | | 著者 | Arenz, S, Ramu, H, Gupta, P, Berninghausen, O, Beckmann, R, Vazquez-Laslop, N, Mankin, A.S, Wilson, D.N. | | 登録日 | 2013-10-23 | | 公開日 | 2014-03-26 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide.
Nat Commun, 5, 2014
|
|
3J7Z
 
 | | Structure of the E. coli 50S subunit with ErmCL nascent chain | | 分子名称: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | 著者 | Arenz, S, Meydan, S, Starosta, A.L, Berninghausen, O, Beckmann, R, Vazquez-Laslop, N, Wilson, D.N. | | 登録日 | 2014-08-27 | | 公開日 | 2014-10-22 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Drug Sensing by the Ribosome Induces Translational Arrest via Active Site Perturbation.
Mol.Cell, 56, 2014
|
|
1Y1N
 
 | | Identification of SH3 motif in M. Tuberculosis methionine aminopeptidase suggests a mode of interaction with the ribosome | | 分子名称: | Methionine aminopeptidase 1B, POTASSIUM ION | | 著者 | Addlagatta, A, Quillin, M.L, Omotoso, O, Liu, J.O, Matthews, B.W. | | 登録日 | 2004-11-18 | | 公開日 | 2005-05-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Identification of an SH3-Binding Motif in a New Class of Methionine Aminopeptidases from Mycobacterium tuberculosis Suggests a Mode of Interaction with the Ribosome
Biochemistry, 44, 2005
|
|
4V65
 
 | | Structure of the E. coli ribosome in the Pre-accommodation state | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Devkota, B, Caulfield, T.R, Tan, R.-Z, Harvey, S.C. | | 登録日 | 2008-08-03 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | The Structure of the E. coli Ribosome Before and After Accommodation: Implications for Proofreading
To be Published
|
|
8RQ0
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation II | | 分子名称: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 88, ... | | 著者 | Lauer, S, Nikolay, R, Spahn, C. | | 登録日 | 2024-01-17 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
8RPY
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api137 | | 分子名称: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 137, ... | | 著者 | Lauer, S, Nikolay, R, Spahn, C. | | 登録日 | 2024-01-17 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
8RQ2
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation III | | 分子名称: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 88, ... | | 著者 | Lauer, S, Nikolay, R, Spahn, C. | | 登録日 | 2024-01-17 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
8RPZ
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation I | | 分子名称: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 88, ... | | 著者 | Lauer, S, Nikolay, R, Spahn, C. | | 登録日 | 2024-01-17 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
6IY7
 
 | |
