9CMH
 
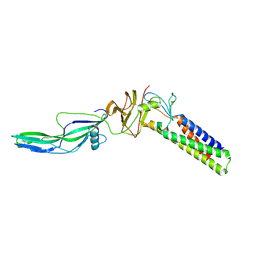 | |
9EM6
 
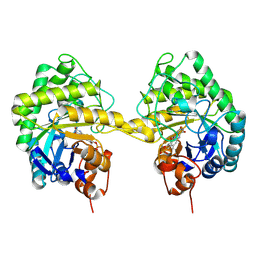 | |
9EQM
 
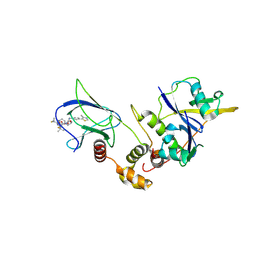 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-21 | | 分子名称: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[trans-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | 著者 | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | 登録日 | 2024-03-21 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
9EYW
 
 | | Human PRMT5 in complex with AZ compound 21 | | 分子名称: | (3~{S})-2-[(5-azanyl-1~{H}-pyrrolo[3,2-b]pyridin-2-yl)methyl]-6-fluoranyl-1'-[(4-fluorophenyl)methyl]spiro[isoindole-3,3'-pyrrolidine]-1,2'-dione, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | 著者 | Debreczeni, J. | | 登録日 | 2024-04-09 | | 公開日 | 2024-08-14 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
9CBS
 
 | |
9FST
 
 | | Yeast 20S proteasome with human beta1i (1-51) in complex with epoxyketone inhibitor LU-001i | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-21 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
To Be Published
|
|
9ATN
 
 | |
9BB6
 
 | |
9HVP
 
 | | Design, activity and 2.8 Angstroms crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease | | 分子名称: | HIV-1 Protease, benzyl [(1R,4S,6S,9R)-4,6-dibenzyl-5-hydroxy-1,9-bis(1-methylethyl)-2,8,11-trioxo-13-phenyl-12-oxa-3,7,10-triazatridec-1-yl]carbamate | | 著者 | Neidhart, D.J, Erickson, J. | | 登録日 | 1990-11-06 | | 公開日 | 1992-04-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Design, activity, and 2.8 A crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease.
Science, 249, 1990
|
|
9EVQ
 
 | |
9FBF
 
 | | VDR complex with UG-481 | | 分子名称: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-7~{a}-methyl-1-[(1~{S})-5-methyl-1-[(1~{R},2~{R})-2-(3-methyl-3-oxidanyl-butyl)cyclopropyl]-5-oxidanyl-hexyl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Rochel, N. | | 登録日 | 2024-05-13 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of New Type of Gemini Analogues with a Cyclopropane Moiety in Their Side Chain.
J.Med.Chem., 67, 2024
|
|
9FCE
 
 | | BelI in complex with SAM from Streptomyces sp. UCK14 | | 分子名称: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | 著者 | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | 登録日 | 2024-05-15 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
9CL9
 
 | |
9BJ0
 
 | |
9BB5
 
 | |
9FCS
 
 | | CysG(N-16)-H122N mutant in complex with SAH from Kitasatospora cystarginea | | 分子名称: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | 著者 | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | 登録日 | 2024-05-15 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
9B6E
 
 | | Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 | | 分子名称: | 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 8 | | 著者 | Yin, Y, Park, C.-G, Zhang, F, Fedor, J, Feng, S, Suo, Y, Im, W, Lee, S.-Y. | | 登録日 | 2024-03-25 | | 公開日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8.
Sci Adv, 10, 2024
|
|
9CV0
 
 | |
9AZJ
 
 | |
7AR5
 
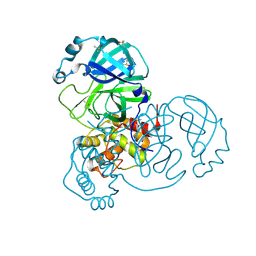 | | Structure of apo SARS-CoV-2 Main Protease with small beta angle, space group C2. | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-10-23 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
9AZB
 
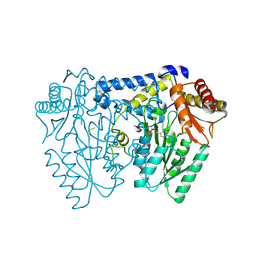 | | Crystal structure of LolTv5 | | 分子名称: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Gao, J, Hai, Y. | | 登録日 | 2024-03-11 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
9EU6
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 23j | | 分子名称: | (1,5-dimethylpyrazol-4-yl)methyl (2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | 著者 | Meyners, C, Buffa, V, Hausch, F. | | 登録日 | 2024-03-27 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-09-11 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | 1,4-Pyrazolyl-Containing SAFit-Analogues are Selective FKBP51 Inhibitors With Improved Ligand Efficiency and Drug-Like Profile.
Chemmedchem, 19, 2024
|
|
9AUM
 
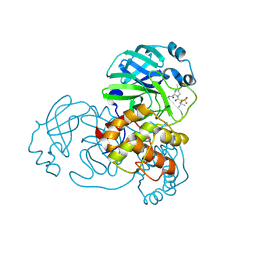 | | Structure of SARS-CoV-2 Mpro mutant (T21I,S144A,T304I) in complex with Nirmatrelvir (PF-07321332) | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2024-02-29 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.539 Å) | | 主引用文献 | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
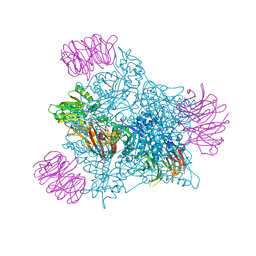
9BRT
 
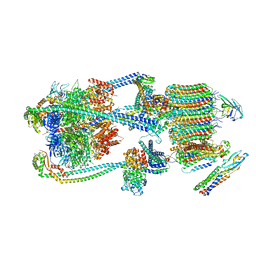 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | 分子名称: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | 著者 | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | 登録日 | 2024-05-11 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
7ASS
 
 | |
