6MWF
 
 | |
7OGI
 
 | | Human Cyclophilin D in complex with N-(5-ethyl-4-oxo-1,2,3,4,5,6-hexahydro-1,5-benzodiazocin-8-yl)-7methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide | | 分子名称: | DIMETHYL SULFOXIDE, N-(5-ethyl-4-oxidanylidene-1,2,3,6-tetrahydro-1,5-benzodiazocin-8-yl)-7-methyl-2-oxidanylidene-1H-pyrazolo[1,5-a]pyrimidine-6-carboxamide, Peptidyl-prolyl cis-trans isomerase F, ... | | 著者 | Silva, D.O, Graedler, U, Bandeiras, T.M. | | 登録日 | 2021-05-06 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.01 Å) | | 主引用文献 | Human Cyclophilin D in complex with N-(5-ethyl-4-oxo-1,2,3,4,5,6-hexahydro-1,5-benzodiazocin-8-yl)-7methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide
To Be Published
|
|
6N9E
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic CC-Pmn and bound to mRNA and P-site tRNA at 3.7A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | 著者 | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | 登録日 | 2018-12-03 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
6NMO
 
 | |
8GQP
 
 | | Complex of D-protein binder D-19437 and L-target L-Pep-1 | | 分子名称: | D-binder, L-pep1 | | 著者 | Liang, M.F, Li, S.C, Wang, T.Y, Liu, L, Lu, P.L. | | 登録日 | 2022-08-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-09-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
6ZJX
 
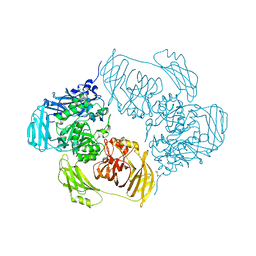 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with saccharose | | 分子名称: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
6X3B
 
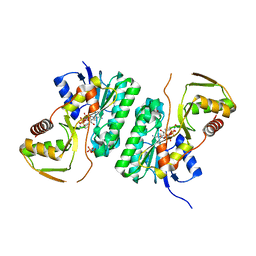 | |
6ZJT
 
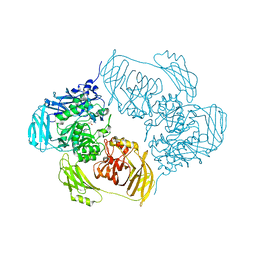 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactulose | | 分子名称: | ACETATE ION, Beta-galactosidase, SODIUM ION, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
6OI9
 
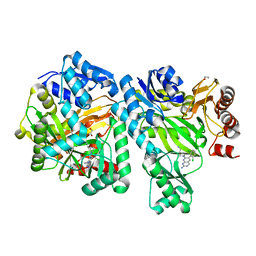 | | Crystal Structure of E. coli Biotin Carboxylase Complexed with 7-[3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine | | 分子名称: | 1,2-ETHANEDIOL, 7-[(3S)-3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | 著者 | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | 登録日 | 2019-04-09 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
6OJU
 
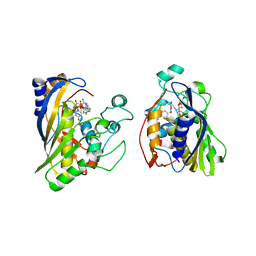 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-D-glutamic acid | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N-{4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}-D-glutamic acid, Thymidylate synthase,Thymidylate synthase | | 著者 | Czyzyk, D.J, Anderson, K.S, Valhondo, M, Jorgensen, W.L. | | 登録日 | 2019-04-12 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.884 Å) | | 主引用文献 | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
6N9F
 
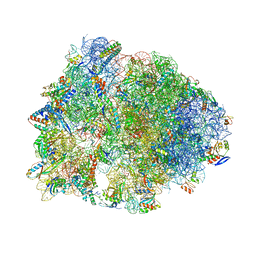 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic ACCA-DPhe and bound to mRNA and P-site tRNA at 3.7A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | 著者 | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | 登録日 | 2018-12-03 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
6MYV
 
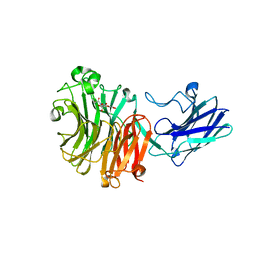 | | Sialidase26 co-crystallized with DANA-Gc | | 分子名称: | 2,6-anhydro-3,5-dideoxy-5-[(hydroxyacetyl)amino]-D-glycero-L-altro-non-2-enonic acid, Sialidase26 | | 著者 | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Huang, J, Siegel, D, Chang, G, Varki, A, Zengler, K. | | 登録日 | 2018-11-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
7Q3S
 
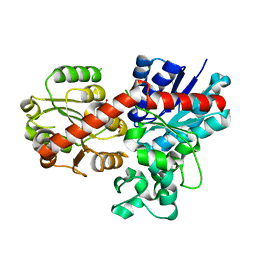 | |
6OI8
 
 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with 7-((1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl)-6-(2-chloro-6-(pyridin-3-yl)phenyl)pyrido[2,3-d]pyrimidin-2-amine | | 分子名称: | 1,2-ETHANEDIOL, 7-[(1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl]-6-[2-chloro-6-(pyridin-3-yl)phenyl]pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | 著者 | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | 登録日 | 2019-04-09 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
8FFF
 
 | |
6ZJU
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with saccharose | | 分子名称: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
6ZJR
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with lactulose | | 分子名称: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
6OTU
 
 | |
8A9M
 
 | |
6ZIS
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer with bound high affinity inhibitor | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, N-{(1S)-5-amino-1-[(4-pyridin-4-ylpiperazin-1-yl)carbonyl]pentyl}-3,5-dibromo-Nalpha-{[4-(2-oxo-1,4-dihydroquinazolin-3 (2H)-yl)piperidin-1-yl]carbonyl}-D-tyrosinamide, TETRAETHYLENE GLYCOL, ... | | 著者 | Southall, S.M. | | 登録日 | 2020-06-26 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structure-Based Drug Discovery ofN-((R)-3-(7-Methyl-1H-indazol-5-yl)-1-oxo-1-(((S)-1-oxo-3-(piperidin-4-yl)-1-(4-(pyridin-4-yl)piperazin-1-yl)propan-2-yl)amino)propan-2-yl)-2'-oxo-1',2'-dihydrospiro[piperidine-4,4'-pyrido[2,3-d][1,3]oxazine]-1-carboxamide (HTL22562): A Calcitonin Gene-Related Peptide Receptor Antagonist for Acute Treatment of Migraine.
J.Med.Chem., 63, 2020
|
|
6ZHO
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer with bound high affinity inhibitor | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | 著者 | Southall, S.M. | | 登録日 | 2020-06-23 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-Based Drug Discovery ofN-((R)-3-(7-Methyl-1H-indazol-5-yl)-1-oxo-1-(((S)-1-oxo-3-(piperidin-4-yl)-1-(4-(pyridin-4-yl)piperazin-1-yl)propan-2-yl)amino)propan-2-yl)-2'-oxo-1',2'-dihydrospiro[piperidine-4,4'-pyrido[2,3-d][1,3]oxazine]-1-carboxamide (HTL22562): A Calcitonin Gene-Related Peptide Receptor Antagonist for Acute Treatment of Migraine.
J.Med.Chem., 63, 2020
|
|
6X9B
 
 | |
8BMW
 
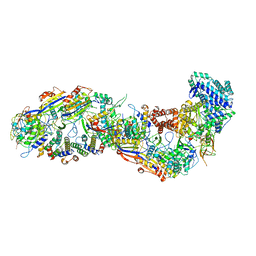 | | SsoCsm | | 分子名称: | CRISPR-associated Cas7 paralog (Type III-D), CRISPR-associated protein Cas10 (Type III-D), CRISPR-associated protein Cas5 (Type III-D), ... | | 著者 | Spagnolo, L, White, M.F. | | 登録日 | 2022-11-11 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-03-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the Saccharolobus solfataricus type III-D CRISPR effector.
Curr Res Struct Biol, 5, 2023
|
|
6X9A
 
 | |
6NZ8
 
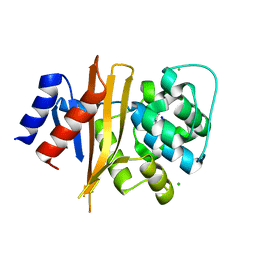 | | Structure of carbamylated apo OXA-231 carbapenemase | | 分子名称: | Beta-lactamase OXA-231, CHLORIDE ION, SODIUM ION | | 著者 | Favaro, D.C, Llontop, E.E, Vasconcelos, F.N, Antunes, V.U, Farah, S.C, Lincopan, N. | | 登録日 | 2019-02-13 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Importance of the beta 5-beta 6 Loop for the Structure, Catalytic Efficiency, and Stability of Carbapenem-Hydrolyzing Class D beta-Lactamase Subfamily OXA-143.
Biochemistry, 58, 2019
|
|
